GPT-Enhanced Hierarchical Deep Learning Model for Automated ICD Coding
Volume 9, Issue 4, Page No 21-34, 2024
Author’s Name: Joshua Carberry, Haiping Xu
View Affiliations
Computer and Information Science Department, University of Massachusetts Dartmouth, Dartmouth, MA 02747, USA
a)whom correspondence should be addressed. E-mail: hxu@umassd.edu
Adv. Sci. Technol. Eng. Syst. J. 9(4), 21-34(2024); ![]() DOI: 10.25046/aj090404
DOI: 10.25046/aj090404
Keywords: ICD coding, Deep learning, GPT-4 prompt, Fine-grained data point, Hierarchical classification
Export Citations
In healthcare, accurate communication is critical, and medical coding, especially coding using the ICD (International Classification of Diseases) standards, plays a vital role in achieving this accuracy. Traditionally, ICD coding has been a time-consuming manual process performed by trained professionals, involving the assignment of codes to patient records, such as doctor’s notes. In this paper, we present an automated ICD coding approach using deep learning models and demonstrate the feasibility and effectiveness of the approach across subsets of ICD codes. The proposed method employs a fine-grained approach that individually predicts the appropriate medical code for each diagnosis. In order to utilize sufficient evidence to enhance the classification capabilities of our deep leaning models, we integrate GPT-4 to extract semantically related sentences for each diagnosis from doctor’s notes. Furthermore, we introduce a hierarchical classifier to handle the large label space and complex classification inherent in the ICD coding task. This hierarchical approach decomposes the ICD coding task into smaller, more manageable subclassification tasks, thereby improving tractability and addressing the challenges posed by the high number of unique labels associated with ICD coding.
Received: 22 April 2024, Revised: 01 July 2024, Accepted: 04 July 2024, Published Online: 18 July 2024
1. Introduction
In healthcare, both structured and unstructured information is recorded and stored to ensure that all relevant healthcare processes and patient observations are properly documented. As a key element of the structured data associated with a specific hospital visit, medical codes, such as those in the ICD (International Classification of Diseases) standards [1], are used to accurately describe the visit and indicate the treatment and diagnoses of the patient. ICD coding has long been a labor-intensive manual process performed by trained professionals, requiring meticulous coding of patient records, including doctor’s notes. In this paper, we examine how medical codes can be automatically assigned to reflect the different medical diagnoses a patient may receive during a hospital visit. Due to the high complexity of healthcare and medicine, there are thousands of unique ICD codes reflecting a myriad of diagnoses. Among the globally prevalent standards currently used in healthcare and healthcare finance, the ICD codes assigned to a particular hospital visit are highly relevant to the patient’s healthcare. This code assignment allows healthcare and finance professionals to accurately communicate hospital visit information and avoid misunderstandings and inaccuracies in billing and treatment [2], [3], [4]. Therefore, medical coding, the process of assigning the appropriate ICD codes for a specific hospital visit, is an important step in healthcare.
Typically, doctors write comprehensive notes that contain important information related to a patient’s visit. These notes, while containing crucial observations and diagnoses associated with the visit, are largely written in unstructured natural language. That is, these notes are kept in a traditional note-taking style, which is not conducive to communication when compared to highly specific and universally recognized medical codes. Professionals known as medical coders are employed to process the unstructured doctor’s notes into structured lists of medical codes, complete documentation, and improve record keeping and communication within the healthcare ecosystem. However, this task is non-trivial due to the complexity of healthcare, which involves dealing with countless diagnoses, many of which are highly similar and easily confused. Introducing automation into the medical coding process could enhance human performance and help allocate resources to more critical aspects of healthcare. Nonetheless, ICD coding automation faces several challenges [2]. As a classification task, ICD coding requires assigning a unique label to each relevant diagnosis. In practical scenarios, dozens of unique codes may be necessary to describe a particular visit, and appropriate codes must be selected from potentially similar codes that could lead to confusion. In addition to the challenges of the classification task itself, doctor’s notes as input data typically exhibit a number of characteristics that further complicate the coding task. For example, the notes do not have a prescribed uniform structure and are written in natural language, which may vary significantly from doctor to doctor and from institution to institution. The writing may include jargon, abbreviations, and typographical errors such as misspellings. In addition, the notes often span several pages, listing a variety of information that may or may not be useful in assigning ICD codes. This means that there is often a large amount of raw input data, but much of it is not relevant to the specification of a particular ICD code to a diagnosis.
In this study, we present an automated ICD coding method that employs a fine-grained hierarchical procedure to predict the ICD codes to be assigned to a given instance of doctor’s notes. Many existing approaches to automated ICD coding employ the following two main steps: first, a vector representation is generated for the natural language input of the doctor’s notes; second, the vector is fed into a multi-label classifier that outputs all predicted ICD codes at once. We refer to methods that employ this popular strategy as coarse-grained. Unlike the coarse-grained approaches, which attempt to code the entire doctor’s notes document in one shot, producing all predicted codes at once [5], [6], [7], we minimize the complexity of code prediction by performing fine-grained assignments that locate and target diagnoses individually within the notes. Using a fine-grained approach, the various code predictions required to fully code an instance of doctor’s notes can be made separately, thus constituting a series of less complex individual classifications [8]. To further support the classification of a given diagnosis, we use GPT (Generative Pre-trained Transformer) to derive related concepts for a diagnosis and identify sentences in doctor’s notes that are semantically related to the diagnosis. The diagnosis is then combined with the related sentences to form a fine-grained data point that is now ready for classification. Since the classifier is responsible for classifying only one diagnosis at a time, the complexity of classification is reduced compared to classifying all diagnoses at once with multiple labels. Furthermore, each fine-grained data point is a human-understandable footprint that can be reviewed to determine the evidence used to arrive at the prediction for a particular ICD code. The proposed approach incorporates a hierarchical classifier that can further decompose the classification task of a single diagnosis into multiple steps or subclassifications. For example, the first subclassification can identify the disease family, and subsequent subclassifications can become more and more specific until they reach the ICD code prediction. Furthermore, the design of the hierarchical classifier is analogous to human decision-making and ensures a higher level of understandability and explainability for users in healthcare. The main contributions and novelties of the paper are summarized as follows:
- Implemented a fine-grained ICD coding approach that predicts one ICD code at a time, thus limiting the complexity of classification while improving human comprehensibility.
- Demonstrated the feasibility and effectiveness of generative large language model (LLM) GPT-4 for sentence extraction, which greatly improves the performance of downstream ICD code classification.
- Introduced a modular hierarchical approach that leverages existing ICD code organization to enable high performance of automated coding when many unique ICD codes must be considered and improve the human comprehensibility of the classification results.
The rest of the paper is organized as follows. Section 2 discusses related work. Section 3 presents the fine-grained ICD coding approach and deep learning architectures used. Section 4 discusses the GPT-powered related sentence extraction technique used in the fine-grained approach. Section 5 details the automated hierarchical classification approach using deep learning. Section 6 presents a case study and the analysis results. Section 7 concludes the paper and mentions future work.
2. Related Work
Prior to the popularization of public datasets such as Medical Information Mart for Intensive Care III (MIMIC-III) [9], researchers achieved automated medical coding primarily through the use of rule-based systems. These systems can leverage expert knowledge and reduce the need for large-scale training data. In [3], the authors introduced a rule-based system that assigns ICD codes by analyzing sentence elements in radiology reports. The system uses production rules that incorporate domain knowledge and simple logic to draw conclusions about the ICD codes that may be indicated by the reports. The increasing volumes of electronic healthcare records (EHR) over time have allowed the application of methods that require larger amounts of training data. In [4], the authors developed a method to automatically generate a set of rules from radiology report data to predict ICD codes. The automatically generated rules were found to be similarly effective to rule sets handmade by domain experts, which is an encouraging sign for medical coding automation. In [5], the authors introduced one of the first machine learning approaches to automated medical coding. They applied Naïve Bayes-based classifier and established the viability of machine learning approaches for the medical coding task.
More recently, the release of large-scale EHR datasets has enabled the use of deep learning models in automated medical coding. This increased data availability has motivated researchers to explore more resource-intensive methods. One generalized two-step deep learning approach has been successful in automated ICD coding. In this generic approach, the language in a doctor’s notes document is first vectorized, and then the entire document is used as input by a multi-label classifier that predicts ICD codes from the resulting vector. In [6], the authors introduced a method that vectorizes the entire document, which is then passed to a convolutional neural network that outputs the predicted ICD code. In [10], the authors achieved high performance in the ICD coding task by exploring various options for the initial vectorization step and using a BERT-based deep learning classifier for final classification. However, the large amount of text in doctor’s notes poses a unique challenge, as the large input can confuse a deep learning classifier, especially if it is biased toward a limited portion of the input. To address this challenge, some research efforts have focused on developing classifiers that can build more complex representations at multiple levels of analysis, aiming to understand doctor’s notes in terms of overall conceptual or even sentence-level complexity [6], [7], [8]. Other research efforts have addressed this problem by introducing a labeling attention mechanism in deep learning methods to produce different document interpretations for each unique label [11], [12], [13]. While these methods demonstrated effectiveness and enhanced classification performance, they also come with limitations. In particular, these methods employ a two-step strategy known as the coarse-grained approach. In other words, given a report or doctor’s notes document, they treat the document as a single input and then predict all associated ICD codes together. Consequently, these methods confront a more complicated classification challenge, where all labels must be predicted from a single classifier input, and potentially output results that can be difficult for users to interpret. In contrast, we present a fine-grained ICD coding approach that targets and predicts one ICD code at a time. To achieve this, we decompose the automated coding task for doctor’s notes into a series of single-label classifications. This approach reduces the complexity of the medical coding process compared to the multi-label classification method. More specifically, our approach starts with a diagnosis from doctor’s notes and then uses GPT-enhanced sentence extraction to identify the sentences that are semantically related to the diagnosis. This information is grouped into a fine-grained data point for ICD code prediction, which can be examined to reveal evidence used by the classifier for medical coding, thereby improving the explainability and confidence of predictions.
A number of existing studies have exploited the inherent relationships between labels, using hierarchical classifiers to make predictions through a series of classification steps. Hierarchical classifiers are characterized by specially designed subcomponents to handle the various decisions required to achieve a complete classification. These involved decisions are arranged into a hierarchical tree with each node representing a decision. One way to implement the hierarchy is to assign a separate subclassifier to each node of the hierarchy to process the decisions, which is known as the per-node local classifier approach. In this approach, the classifiers required for each decision or subclassification can be specially designed for their particular tasks, and the complexity of each subclassification can be much lower compared to the single step in a non-hierarchical classification approach. The per-node local classifier approach has been used with success in a variety of classification tasks. In [14], the authors used a local classifier per node hierarchy for the galaxy morphology classification task. Their approach used an established taxonomy of galaxy morphologies to design the local classifier hierarchy. In [15], the authors proposed a local classifier-based solution for genomic data classification. Their approach applied multi-label annotation of examples along several paths of the hierarchy and the results showed that deeper and more detailed hierarchies could produce better results. In [16], the authors used a local classifier hierarchy for the task of protein function classification. In their approach, the physical and chemical properties of proteins were used to predict their function in an organism. In [17], the authors employed local classifiers to classify natural language documents by topic. Their approach exploited the hierarchical relationships between related topics and used these relationships to design effective local classifier hierarchies. In [18], the authors studied the design of local classifiers at a higher level and investigated the impact of hierarchical design on classification performance. Their results showed that their hierarchical classification approach could significantly improve classification performance, but the performance gains would depend heavily on a good hierarchical design and training parameters. Unlike the above approaches, our approach is to design a classifier hierarchy by using a taxonomy of ICD codes to enable the automated coding of diagnoses. In our hierarchical approach, we first classify the disease type or family for a given diagnosis, and then attempt to classify specific ICD codes using subclassifiers in a hierarchical tree.
There have also been some efforts to implement hierarchical components in a single classifier, which is known as a global hierarchical classifier. In [19], the authors compared the global Naïve Bayesian classification method with local methods and found that the global Naïve Bayesian classification method has a performance advantage in the protein function classification task. In [20], the authors tackled the face recognition task by combining several techniques, including global hierarchical classification. Their models included a convolutional component consisting of layers that could produce successively more complex classification features. Like hierarchical classification with local classifiers, using a global hierarchical classifier allows designers to address specific aspects of classification through hierarchical components. The resulting global classifier is highly cohesive and easy to train and apply. However, a global classifier lacks the modularity provided by the individuality and separability of local classifiers. Therefore, a global classifier can be difficult to design and it is not possible to train their hierarchical components in a highly specific manner to optimize their performance. In contrast, the modular design in our per-node local classifier approach allows local classifiers to be reused, repurposed, or rearranged in the hierarchy or other hierarchies without the need for extensive retraining. Depending on the nature of the classification task, efficiency can also be improved by parallelizing the training of multiple local classifiers. In addition, the use of local classifiers in a hierarchical architecture allows for more direct decision tracking through the explicit hierarchical decision tree, whereas the hierarchical components and decisions of a global classifier reside in the black box of a single classifier, and thus interpreting the behavior of the global classifier may be more difficult.
One of the major performance bottlenecks in ICD coding is extreme labeling bias. That is, some ICD codes are so frequent that thousands of unique examples may appear in a given dataset, while other codes may represent rare diagnoses with only a few examples. Typical deep learning methods require many examples to learn a given label, and are therefore particularly vulnerable to label bias introduced by ICD codes. Several recent studies have specifically addressed the problem of label bias in an attempt to directly address this key challenge. In [21], the authors used a debiasing method that first statistically analyzes the model’s performance to detect bias. Once quantified, the model’s bias for each class is used to calculate a debiasing factor, which is utilized to adjust the confidence of the model’s output for each class before deciding on the final prediction. In addition to deep learning models, some researchers used fuzzy logic and string matching techniques to improve the performance of few-shot and zero-shot ICD coding [22]. After initially identifying the ICD code category through a deep learning classifier, fuzzy string matching was used to compute Levenshtein distances between sentences in the doctor’s notes and the various ICD codes included in the predicted category from which the final code predictions were selected. In [23], the authors explored the use of transfer learning in a related unsupervised learning task to provide additional learning data for the deep learning classifier. In their approach, the classifier is trained not only on labeled examples that can be very scarce especially for underrepresented ICD codes, but also on completely unlabeled clinical texts using token-level similarity. In this paper, we take a hierarchical approach to the ICD coding problem, building labeling relationships (between different ICD codes) into the model architecture. This predefined hierarchical structure leverages the existing knowledge about labels and supports label classification, even if few examples can be trained. However, beyond this hierarchical structure and the application of class weights during training, the problem of label bias is not directly addressed. Therefore, the above strategies for mitigating label bias can be used as a complement to our approach in future research.
While LLMs and generative AI are best known for their generative capabilities through chatbot applications such as ChatGPT and Microsoft Copilot, they can also be used for more straightforward tasks like text classification [24], [25]. Most LLMs have a large internal network whose output is passed through a specially trained “head” to produce the final desired output. One way to adapt an LLM to perform classification is to replace the generative head with a classification head. The network can then be trained holistically to learn the classification task and fine-tuned using the existing knowledge to be applied to that task [26]. This approach has been used in Google’s BERT LLM and its many variants [27]. In [28], the authors applied several BERT-based models to influential text classification benchmarks and achieved state-of-the-art results. Some of the benchmarks involved include topic classification, sentiment classification, and goods and services identification. Similar to these methods, we use BERT-based classifiers with a multiclass classification head to perform classification tasks. In addition to being used directly for classification tasks, the generative capabilities of LLM can also be used to augment or enrich the input data points for other classification methods. In [29], the authors introduced GPT3Mix, a method that augments training data by using the generative ability of GPT-3. In GPT3Mix, GPT-3 combined multiple training examples to generate a hybrid synthetic training example, and downstream classifiers trained on GPT3Mix-enhanced data points showed significant improvement over the baseline model. In [30], the authors utilized LLM-based text augmentation to improve classification performance for grant proposal research topics. Their approach enhances imbalanced training data by targeting underrepresented classes and generating new training data points to populate them. In contrast to these methods, our medical coding approach enriches data points indirectly by using generative language models. Based on an initial diagnosis concept, we prompt the model for a set of semantically related terms. We then use these related terms to mine related sentences from the free text of doctor’s notes and combine them with the diagnosis to generate an enriched fine-grained data point to improve training and classification performance.
3. Fine-Grained ICD Coding Using Deep Learning
3.1. A Novel Approach for Automated ICD Coding
During a hospital visit, healthcare professionals collect and record various data about the patient. One key record comes in the form of doctor’s notes, which are text-based records generated and maintained by hospital staff. Doctor’s notes typically cover the entire healthcare process and may involve anything from medical measurements and observations to patient medical histories and miscellaneous comments. Some parts of doctor’s notes are loosely structured, presenting information in bulleted or numbered lists; while others can be unstructured, presented in common sentences with agrammatic or misspelled language. Figure 1 shows a text snippet from a randomly sampled example of doctor’s notes.

The free-text format of doctor’s notes allow
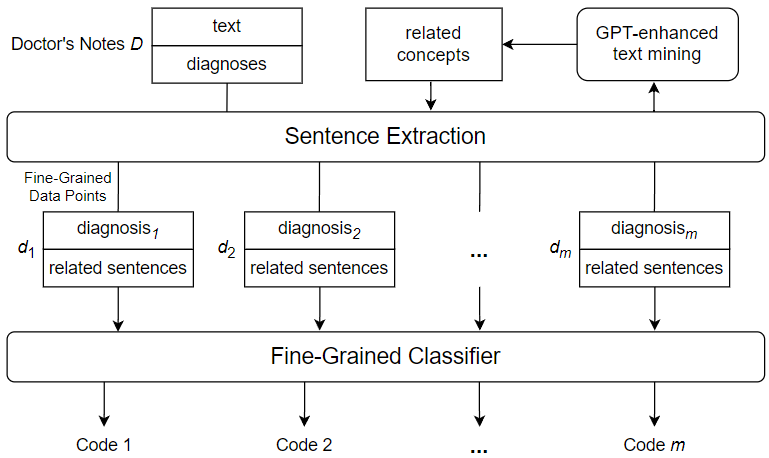
s for flexibility and convenience in covering a wide range of information about the patient and their hospitalization, but this flexibility comes at a price. Because of the free-form nature of doctor’s notes, much of the data they encode is not sufficiently structured and organized to be used effectively. For example, a doctor may have to flip through pages of irrelevant patient history to find important details related to a specific diagnosis; and an external institution such as an insurance company may not be able to recognize information related to billing due to the peculiarities of the way it is written. For this reason, doctor’s notes must be annotated with a set of highly specific codes that show the exact diagnoses and course of treatment. This allows other healthcare professionals and external entities to quickly and directly assess critical information that has previously been obscured by the difficulties associated with doctor’s notes. The ICD international standard provides a robust and extensive set of medical codes used to identify a myriad of disease diagnoses in healthcare. A version of the standard, ICD-9, was widely used in modern healthcare, leading to the release of a number of datasets coded using the standard. Recently, the newer ICD-10 has been accepted and frequently used by hospitals in the United States and some other countries. There are several accessible datasets using the ICD-10 standard, which are used for a variety of machine learning tasks, including automated ICD coding. The latest standard, ICD-11, has not yet been widely adopted, thus the available data using this standard are limited. However, ICD-11, like its predecessors, greatly expands the code bases offered by the previous standards and provides an important research motivation for the topic of automated coding using a large number of unique ICD codes. Despite the importance of ICD coding in healthcare, identifying and assigning the appropriate codes for a given instance of doctor’s notes has been a non-trivial task. To maintain manageable classification complexity and enhance ICD coding accuracy, we introduce a novel approach to automated ICD coding of doctor’s notes using GPT-enhanced text mining. Figure 2 shows an overview of the key components and steps of the proposed automated ICD coding approach. As shown in the figure, we use a fine-grained method, which performs ICD code assignment as a series of single-label classifications rather than a single multi-label classification. This means that the classifier used is only responsible for predicting one ICD code from a given input, thus reducing the complexity of the ICD code prediction process. In order to construct the fine-grained data points used to code a given instance of doctor’s notes, a diagnosis needs to be selected from the doctor’s notes and paired with related sentences from the free text of the doctor’s notes. To mine sentences related to a specific diagnosis, we derive a set of diagnosis-related concepts using GPT-enhanced text mining. Subsequently, we search the free text of the doctor’s notes to mine sentences that contain one or more of the derived related concepts. The extracted related sentences are then combined with the diagnosis to form a fine-grained data point, which is fed into the fine-grained classifier to generate an ICD code prediction.
3.2. Fine-Grained ICD Code Assignment
Unlike the fine-grained approach used in this paper, existing methods typically use coarse-grained techniques for ICD coding. These methods perform ICD coding through multi-label classification. That is, given a single input, which is the entire contents of a doctor’s notes instance, the goal is to produce a set of outputs, i.e., all appropriate ICD codes for the doctor’s notes instance. The classification procedure performs only one operation to process the entire doctor’s notes as a whole and outputs the predicted ICD codes accordingly. Figure 3 shows an overview of a typical coarse-grained classification approach.
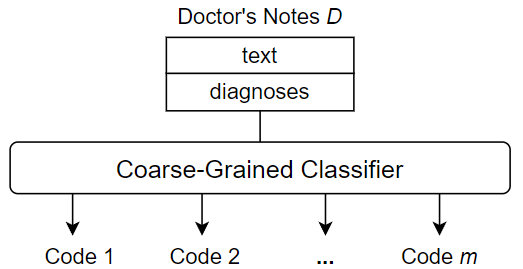
One of the main disadvantages of coarse-grained methods is that they can lead to very complex classifications. First, the classifier has to deal with a large amount of raw data, as the contents of doctor’s notes can be several pages long. Much of this information is not useful for a specific ICD code assignment, and some may be useless for any code assignments, essentially useless noise. Second, a multi-label classification is inherently difficult, especially when the number of unique labels to be assigned is large. The ICD standards currently in use contain thousands of unique codes, and the code base continues to grow as new versions are released. Therefore, large-scale ICD coding is considered to be a difficult classification task with a very large label space. That is, when the number of unique labels in a multi-label classification task is , then there exist unique label combinations. The resulting complexity must be captured by the training data and be tractable for the classifier architecture, which becomes increasingly challenging. In contrast, we adopt a fine-grained approach to ICD code assignment that seeks to avoid the difficulties associated with a coarse-grained strategy. The key to the fine-grained approach is to subdivide the ICD code assignment process down to the individual code level. In other words, rather than predicting all codes at once, we predict one code at a time from some “starting point”, namely a diagnosis, in the doctor’s notes. This shifts the task from a multi-label classification to single-label classifications, thus limiting the inherent classification complexity. Since we are predicting one code at a time, there is also no need to overwhelm the classifier with the full text of the doctor’s notes, instead using only the text that we believe is relevant to the current code classification. Thus, fine-grained classification not only reduces the complexity of the classification step, but also provides the opportunity to trim or enrich the input data used to predict each individual code in the medical coding process.
To facilitate the division of the ICD code assignment task for a doctor’s notes instance, we can utilize the diagnoses in the doctor’s notes to mark the presence of codes to be identified. A doctor’s notes instance typically contains a section of “Discharge Diagnoses,” which is a delimited list of the most important diagnoses at the time of a given patient’s visit. Figure 4 shows the discharge diagnoses from randomly sampled doctor’s notes.

While the diagnoses provide the initial concepts for assigning codes to the doctor’s notes, they often lack the specific details required to determine the assignment of individual ICD codes. These diagnoses may contain abbreviations, misspellings, or other incompleteness that preclude precise assignment. Therefore, while they cannot be used alone to draw conclusions about the ICD codes needed for a given doctor’s notes instance, they can serve as ideal starting points for fine-grained classifications. For each diagnosis in a discharge diagnoses section, we perform a single-label classification on a different subset of the notes to predict the corresponding code. After assigning a predicted code to each diagnosis, the resulting set constitutes the set of predicted codes for the entire doctor’s notes instance. Let Ffine be a fine-grained classifier defined as a function that outputs an array of confidences for the output classes. Let doctor’s notes D be a 2-tuple (DIAG, FTXT), where DIAG is a list of diagnoses and FTXT is the free text of the doctor’s notes, respectively. The procedure for classifying an instance D of doctor’s notes using a fine-grained classifier Ffine is described in Algorithm 1. As shown in the algorithm, each individual diagnosis in the discharge diagnosis section is combined with a set of semantically related sentences in the free-text of the doctor’s notes to form a fine-grained data point dp. The data point dp is then used as an input to the single-label multiclass ICD code classifier Ffine, which predicts a suitable ICD code. Once all diagnoses in the doctor’s notes have been processed, the generated ICD code set is returned as the predicted code set for the doctor’s notes. Note that in our fine-grained approach, sentences in the doctor’s notes are appropriately ignored if they are not relevant for the classification of a diagnosis. Therefore, our approach utilizes targeted and useful information for each prediction, enabling the classifier to predict relevant ICD codes more accurately. In addition, since the amount of raw text in the doctor’s notes may pose a problem for a classifier architecture that is limited by the size of the input, sentence extraction provides yet another key benefit. By removing irrelevant information, we also limit the average length of the data points, thereby expanding the scope of applicable classifiers and training techniques, which were previously limited by potentially high data volumes. A key consideration when using a fine-grained approach is how to identify the subset of doctor’s notes that are relevant and useful for the classification of a particular diagnosis. Thus, it is critical to develop an effective method for the inclusion of specific text passages in doctor’s notes based on their usefulness.
| Algorithm 1: Automated ICD Code Assignment | |
|
Input: an instance of doctor’s notes dNotes, a single-label multiclass ICD code classifier Ffine Output: a set of m predicted ICD codes codeSet, where m = |dNotes.DIAG| |
|
|
Initialize codes = ∅ for each diagnosis α in dNotes.DIAG: |
|
|
Extract a set of sentences Ψ from dNotes.FTXT related to α Let fine-grained data point dp be (α, Ψ) confidences = Ffine(dp) code = argmax(confidences) codeSet = codeSet ∪ {code} |
|
|
end return codeSet |
|
3.3. Transformer-Based Deep Learning Models
Deep learning models are one of the dominant tools in the field of Natural Language Processing (NLP) [31]. Deep learning utilizes deep neural networks to perform tasks that are often difficult to solve programmatically because they are complex and potentially poorly-defined. Neural networks are computational architectures composed of artificial neurons. As shown in Figure 5, an artificial neuron takes one or more values as inputs and transform these values to generate an output value. The output value is either sent to the next neuron or neurons for further computation or as the final output of the network.
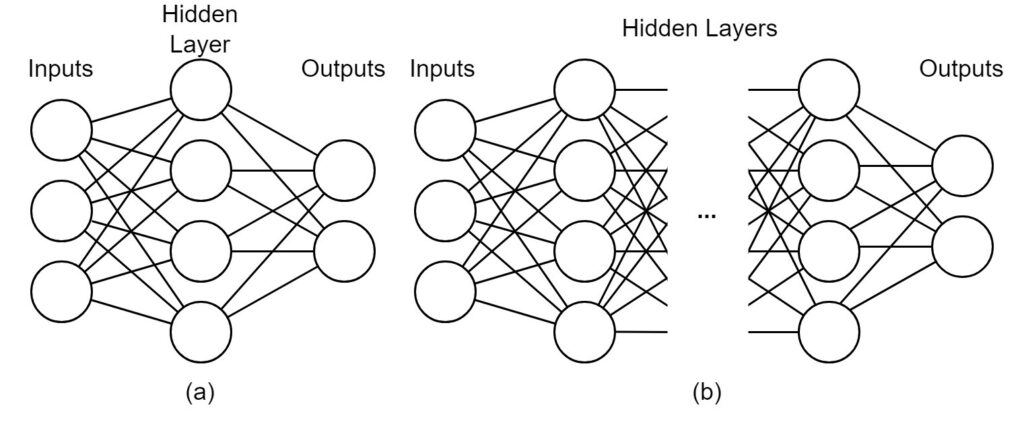
During the passage through the neural network, the input values are transformed according to a set of learned parameters called model weights. These weights are learned during the training phase, which optimizes the weights according to some correctness criteria. For example, a neural network classifier is trained to predict the true class of a given example, which means that the network will be optimized towards correct classification. After training, the network should be able to predict classes with reasonable classification performance even for unseen examples. While traditional neural networks contain only one hidden layer of neurons, limiting the complexity of possible transformations and resulting performance, deep neural networks use any number of hidden layers, meaning that arbitrarily complex architectures can be developed to cope with difficult problems. A comparison between a traditional neural network and a deep neural network is illustrated in Figure 5 (a) and (b).
One deep learning architecture that dominates in NLP is the transformer-based architecture. A transformer-based architecture is a deep neural network consisting of modules called encoders and decoders equipped with self-attention mechanisms. These self-attention mechanisms allow the network to learn and express relationships between individual tokens (e.g., words) in a natural language input sequence. At a high level, transformer-based architectures provide the complexity necessary to capture the way language changes based upon its context. One example is homonymy, where the same pronunciation or spelling has different meanings. For example, “saw” can represent one of two meanings depending on context. Take this sentence for example: “Patient reported he saw black spots in his vision,” where “saw” comes from the verb “see.” In another sentence: “Patient admitted with injuries related to a power saw,” “saw” is referring to a power tool. While a more primitive NLP model may assign the same meaning to both usages of “saw,” the self-attention mechanisms in a transformer-based deep learning network allow the model to distinguish between the two uses, resulting in a more robust and accurate understanding of the language. In essence, self-attention mechanisms allow the model to process each token (e.g., word) in the input sequence considering its relations to the surrounding tokens. When found nearby the word “vision”, “saw” is likely to refer to eyesight. On the other hand, when “power” is located nearby, “saw” is likely to refer to power tools. Other words that may have different meanings include pronouns, i.e., “it,” “they,” “these,” “those,” and other non-specific nouns used to denote other nouns. Whereas a more primitive NLP model may make little use out of such words, self-attention models can decipher the meaning of pronouns and give them the proper treatment. Figure 6 shows how an attention-based model can characterize a pronoun that other models may not understand.
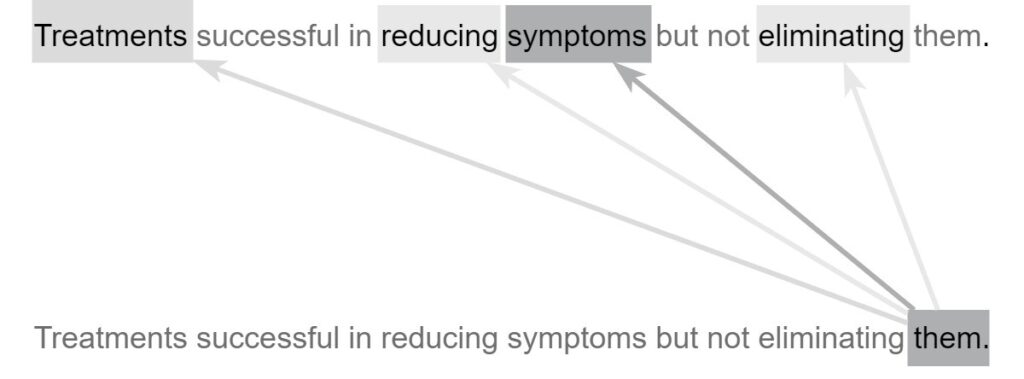
As shown in Figure 6, the word “them” should not be taken at face value. Instead, the attention mechanism uses the surrounding words to modify its meaning to give it a more specific and useful characterization. The word “them” attends most strongly to the word “symptoms” because it is the actual word “them” refers to. It also attends weakly to “treatments,” a word related to “symptoms” and attends slightly to the verbs “reducing” and “eliminating,” both of which act upon the “symptoms” in the sentence. Transformer-based architectures are characterized by the presence of transformer encoders and decoders, which are special architectural modules containing self-attention mechanisms. Encoders are responsible for converting natural language inputs into vectors. These vectors, known as encodings, can be used for a variety of downstream tasks. Decoders work in the opposite direction, taking encoded vectors as input and converting them back into natural language tokens (e.g., words). For example, a question-answer model may first locate the correct answer to a given question in the semantic or meaning space before using decoders to generate the natural-language expression of the answer. In this paper, we explore the automated ICD coding task using the fine-grained approach described in the previous sections. In our approach, a fine-grained data point is formed using a diagnosis and its semantically related sentences. The fine-grained coding task takes one fine-grained data point (natural language input sequence) and outputs one ICD code. To accomplish this task, some essential pre-processing tasks need to be performed, including lowercasing all letters as well as removing specific dates and identifiers as they are not useful for the classification task. Figure 7 shows a fine-grained classifier used to complete the classification step of the fine-grained ICD coding method.
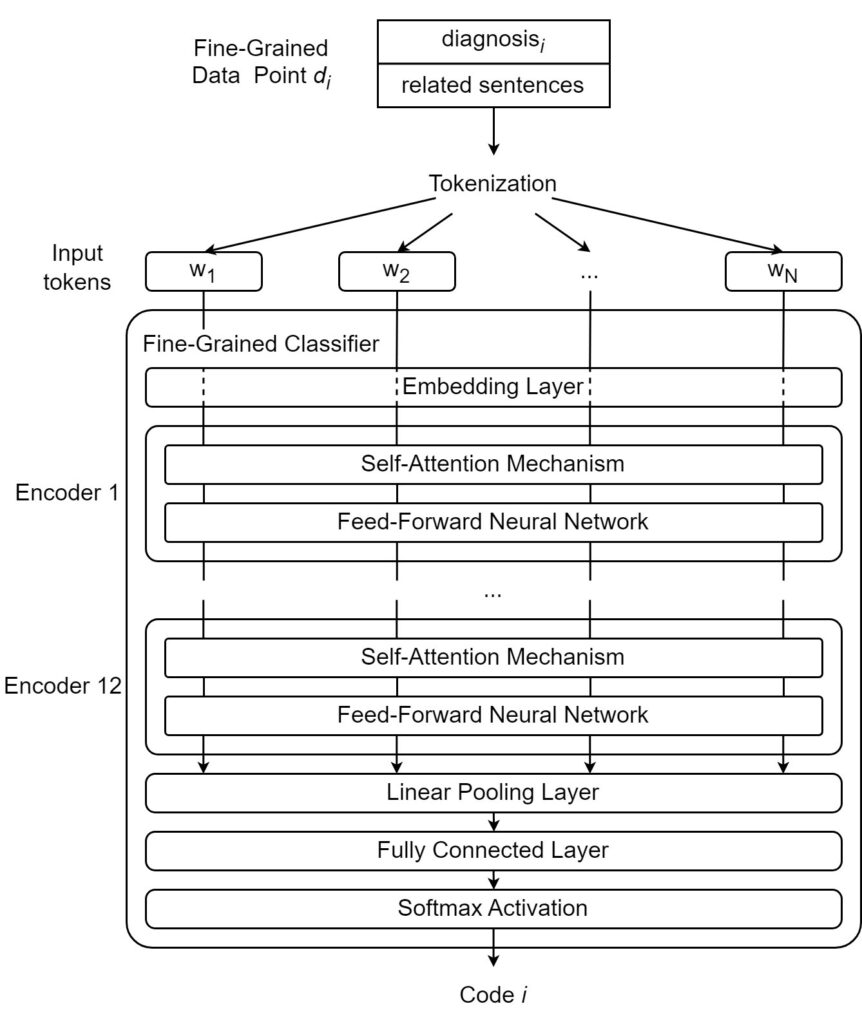
As shown in Figure 7, we tokenize the text by splitting the data point into a sequence of distinct tokens (e.g., words, suffixes). We then employ a fine-grained classifier that performs the following steps: first, tokens are given their initial vector representations or “embeddings” through an embedding layer. Once the original tokens have been converted to embeddings, they are ready to be encoded. In this case, they are fed into the first encoder, where self-attention is applied, and the resulting vectors are fed into a feed-forward neural network that further transforms the vectors. After passing through the neural network, the vectors are released from the first encoder and go to the next encoder. In this paper, we use a BERT-based model that contains a series of 12 encoders. After the 12 encoders have processed the vectors, the fully encoded vector is pooled and passed through a fully connected linear layer. The obtained values are then passed through a Softmax activation layer to output the final prediction.
In addition to the advantages offered by transformer-based architectures, contemporary NLP models such as GPT-4 and BERT benefit greatly from pre-training. That is, they are extensively trained on very large datasets to gain general knowledge of the language and its meaning before being used for a specific application. After pre-training, users can extend or refine the general knowledge of the model for a specific task through further training (i.e., fine-tuning). We use MedBERT [32] as a fine-grained classifier, which is based on Google’s BERT architecture, a transformer-based model. In addition to the general pre-training of BERT on BookCorpus and the English Wikipedia, MedBERT was pre-trained on electronic medical record data, which means that it is particularly well suited for ICD coding. We fine-tuned MedBERT’s pre-training weights using the ICD coding task to maximize its performance. In addition to using MedBERT for classification, we use another transformer-based pre-trained model, GPT-4, to perform the sentence extraction step of the method. In the next section, we present a GPT-powered concept matching approach to support the extraction of semantically related sentences from doctor’s notes to a specific diagnosis.
4. Sentence Extraction Using GPT-4
To extract semantically related sentences from doctor’s notes, we introduce a mechanism for reasoning about the potential relationship between a given sentence and a given diagnosis. Briefly, a sentence can be viewed as a sequence of words referring to a set of concepts. In the most straightforward case, a sentence can directly refer to the concept of a diagnosis. In this case, it is easy to determine that the sentence is related to the diagnosis and can be useful for downstream classification. However, a sentence may also be indirectly related to a diagnosis. For example, a sentence talks about a certain symptom such as “runny nose”; although it never explicitly refers to a diagnosis such as “influenza virus”, it would certainly be related to the diagnosis through the mentioned symptom. Thus, in order to get good coverage of sentences that might be related to a diagnosis, we need to consider not only sentences that talk about the diagnosis, but also sentences that mention other concepts related to the diagnosis. Figure 8 shows the process of sentence extraction using GPT-4. As shown in the figure, the first step in extracting semantically related sentences from doctor’s notes is to generate a set of concepts related to the selected diagnosis. These related concepts can be used in a subsequent search for related sentences in the free text portion of doctor’s notes. Thus, our approach is to identify a set of sentences related to the original diagnosis by one or more related concepts. In previous work, the sentence extraction step was carried out using a set of related concepts generated through an ontology, a knowledge representation that encodes concepts and relations in a directed graph [2]. Ontologies offer a number of advantages for this part of the ICD coding procedure. First, ontologies provide a suitable knowledge base for reasoning about medical concepts and their relationships, which is critical to the sentence extraction step. Furthermore, ontologies can be handcrafted by experts and adapted to the ICD coding process. However, the need for manual design of ontologies can be seen as a weakness, as it incurs development costs and requires expert domain knowledge. In this paper, we present an alternative approach that utilizes the Chat Completions API of GPT-4 to generate a list of related concepts to be used in the sentence extraction step. Unlike ontologies, GPT-4 is a pre-trained LLM that works out-of-the-box; while its use and performance may require expert monitoring and validation, it does not incur the initial cost of ontology design specifically for ICD coding tasks.
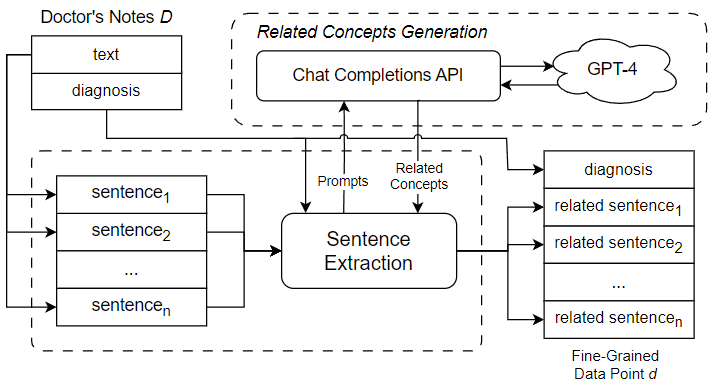
GPT-4, the latest available version of OpenAI’s LLM, was trained on a large text dataset mined from online documents. It has demonstrated capabilities extending beyond standard language processing tasks. In one study, GPT-4 outperformed expected human scores on the official United States Medical Licensing Examination (USMLE), demonstrating its understanding of healthcare and medical concepts [33]. The approach outlined in this paper utilizes the GPT-4’s ability in the healthcare domain to generate a list of related concepts for use in the sentence extraction step. In our approach, we use a diagnosis to generate prompts for GPT-4 designed to elicit a list of related concepts. The prompts are then fed into GPT-4 through its Chat Completions API, which is an interface for the generative features of OpenAI models. Similar to a typical chatroom scenario, the interface accepts an incomplete chat log as an input, from which GPT-4 predicts how the chat will continue by generating the subsequent message.
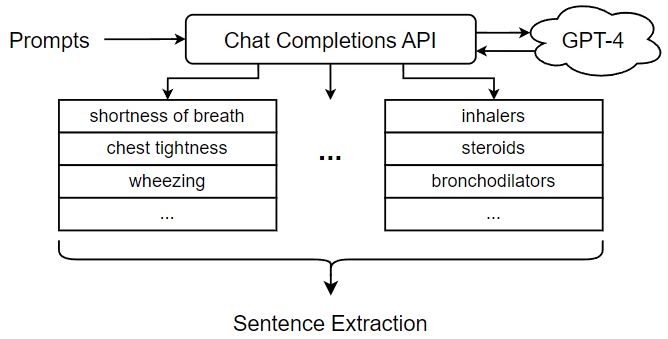
Since the performance of our approach depends on the quality of the related concepts used to perform sentence extraction, we focused a great deal of attention on designing Chat Completions prompts to elicit appropriate behaviors and concepts from GPT-4. Two main factors were considered when designing the prompts. First, some prompt needs to be designed to direct GPT-4 to properly organize the outputs because we need a list of text strings that correspond to the concepts associated with a given diagnosis. For this reason, we want our Chat Completions prompts to elicit text outputs that are listed, bulleted, or numbered so that we can easily separate the different concepts. To this end, we devise a “system” message specifying that GPT-4 should respond with a dashed list. In this way, the text output by GPT-4 can be reliably separated into individual concepts, which we can then use to extract related sentences. In addition to this generic “system” message, we must design multiple prompts, each including a “user message” representing a query for each subset of related concepts. Table 1 shows several examples of user messages used to elicit concepts related to “Asthma”. Another important requirement for the Chat Completions prompt is that it only elicit concepts related to the diagnosis. Therefore, when extracting related sentences from the free text of doctor’s notes using the related concepts, they can provide useful information or context for predicting the corresponding ICD code for the given diagnosis.
Table 1: Examples of Prompts for Deriving Related Concepts to “Asthma.”
| User Message (Prompt) | Related Concepts (Output) |
| “List the different ways a doctor may indicate the diagnosis ‘Asthma’ in healthcare documentation.” | asthmatic condition, chronic asthmatic, reactive airway disease, asthmatic disorder, … |
| “List the treatments associated with the diagnosis ‘Asthma’.” | inhalers, steroids, bronchodilators, leukotriene modifiers, … |
| “List the symptoms associated with the diagnosis ‘Asthma’.” | shortness of breath, chest tightness, wheezing, coughing, … |
| “List the body parts and organs that may be affected by the diagnosis ‘Asthma’.” | lungs, air passages, bronchial tubes, respiratory tract, … |
Since the purpose of sentence extraction is to generate a fine-grained data point for the classification step, it is desirable to derive related concepts and related sentences to support optimal classification results. To this end, we prompt the LLM to generate several classes of concepts related to the diagnosis in different ways (e.g., synonymy, treatment, symptom, etc.). Algorithm 2 describes the detailed steps to extract a set of semantically related sentences Ψ from free text ξ of doctor’s notes dNotes for a given diagnosis α.
| Algorithm 2: Extract Related Sentences for a Given Diagnosis | |||
|
Input: a given diagnosis α, free text ξ of doctor’s notes dNotes. Output: a set of related sentences Ψ |
|||
|
Initialize a set of related concepts Γ_α = { α } Initialize a set of related sentences Ψ = Define a list of prompts Π to elicit related concepts for α Let delimiter be delimiting symbols (e.g., ‘,’, ‘;’, ‘\n’, …) for each prompt in Π: |
|||
|
Receive a response σ from GPT-4 Chat Completions API Derive a set of concepts Γ_ σ by tokenizing σ on delimiter Γ_α = Γ_α ∪ Γ_ σ |
|||
|
end Split the free text ξ into a list of sentences Σ for each sentence β in Σ: |
|||
|
Derive a set of concepts Γ_β mentioned in β if Γ_β ∩ Γ_α ≠ ∅: |
|||
| Ψ = Ψ ∪ { β } | |||
| end | |||
|
end return Ψ |
|||
As shown in Algorithm 2, we first initialize the set of related concepts Γ_α so that it contains the original diagnosis concept α, which serves as the starting point for deriving related concepts. The set of related sentences Ψ is also initialized to an empty set. We then define a list of prompts to elicit concepts related to α and receive responses using GPT-4 Chat Completions API. These responses are tokenized into related concepts and included in Γ_α. After deriving the related concepts using GPT-4, we divide the free text ξ into a list of sentences, tokenizing each sentence to derive the set of concepts discussed in the sentence. If any concept discussed in a sentence also appears in Γ_α, the sentence is considered relevant and is therefore included in Ψ. Finally, Ψ is returned and combined with the diagnosis text to form a fine-grained data point.
5. Automated Hierarchical Classification Using BERT
Traditionally, classification problems are solved by monolithic or “flat” classifiers, which process inputs and make predictions in a linear path through a single classifier or module. Monolithic classifiers are easy to implement and provide good performance in a variety of classification tasks, especially if the number of unique labels is kept reasonable. However, monolithic classification methods are not scalable within a given classifier architecture; as more unique labels are added to the classification task, the complexity required to model the extended label space eventually becomes too great, compromising classification performance. In this paper, we introduce a hierarchical classification approach that predicts the corresponding ICD code for a fine-grained data point through a series of classification steps of increasing specificity. Depending on the design, a hierarchical classifier can first identify the general type of disease and then narrow it down for more precise classification until a specific ICD code classification is derived. In our hierarchical classification approach, additional subclassifiers or classification modules are added, each of which is responsible for a certain step in the overall classification process. We refer to these classification steps as subclassifications. For example, the first subclassification of a given instance of doctor’s notes might be a binary determination of whether it is a respiratory disease or a circulatory disease, while the second subclassification, which is more specific, might be to determine which individual disease to code for. Figure 9 compares a monolithic classification architecture with a hierarchical classification architecture that consists of three subclassifiers A, B and C, where A is the root subclassifier for the hierarchical classification.
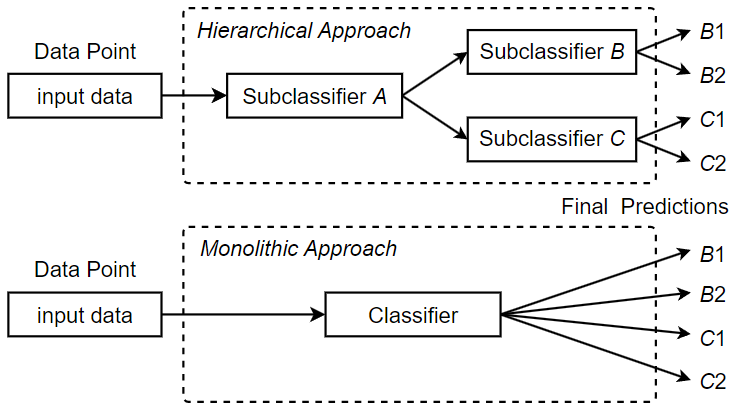
subclassifiers forms a classification hierarchy, where data points flow from the root subclassifier to a leaf subclassifier. The main benefit of using a hierarchy of subclassifiers is that it scales to the large label spaces associated with the ICD coding task. When greater code coverage is required, the classification can be split into individual subclassifications of lower complexity. Each subclassification can then be covered by a separate subclassifier. As long as the subclassifications are sufficiently tractable, classification performance remains high at every step, and the overall performance can be maintained despite the additional complexity introduced by the ever-expanding label space. We define a single-label multiclass subclassifier Fsub as a 2-tuple (FSUB, CHID), where FSUB is the prediction function of the subclassifier, which outputs an array of confidences for the output classes, and CHID is an array of children subclassifiers, or the empty set if the classifier has no further children. A special subclassifier Froot is defined as the root subclassifier for hierarchical classification. Algorithm 3 describes the detailed procedure of hierarchical classification. As shown in the algorithm, using a hierarchical classification approach, a fine-grained data point dp is first processed by the root subclassifier Froot. Based on the confidence level of the output classes, the child subclassifier corresponding to the class with the highest confidence level is selected for further subclassification. This process is repeated until a leaf subclassifier with no child subclassifiers is reached. In this case, the ICD code corresponding to the class predicted by the leaf subclassifier is returned as the matching code for dp.
| Algorithm 3: Hierarchical Classification | |
|
Input: a fine-grained data point dp, the root subclassifier Froot for the hierarchical classification Output: the predicted ICD code code |
|
|
Let Fcurrent be the current single-label multiclass subclassifier Initialize Fcurrent = Froot while Fcurrent : |
|
|
confidences = Fcurrent.FSUB(dp) class = argmax(confidences) if Fcurrent.CHID ≠ then Fcurrent = Fcurrent.CHID[class] else Fcurrent = |
|
|
End Set code to the ICD code corresponding to the predicted class return code |
|
It is worth noting that the hierarchical implementation also allows for a degree of modularity in the classification methodology, which can be used for multiple purposes in ICD auto-coding applications. The introduction of hierarchical organization divides the overall classification into discrete subclassification steps designed by domain experts. These divisions and the resulting subclassifications necessarily have human-understandable meanings. Thus, our hierarchical approach has the advantage that even users unfamiliar with the details of deep learning can have some understanding of the various decisions made by the final classification. In other words, it is possible to examine and study the order of the subclassifications or the decision path that led to the final classification. This enhances the trustability that code assignments are made in a coherent and consistent manner and adds to the overall interpretability and credibility of the method. Finally, the modular design of the hierarchical classification facilitates performance analysis, and in cases where classification is difficult or unclear, users or developers can track the subclassifications involved to gain clarification or identify erroneous features. During training and testing, the errors caused by each subclassifier can be examined on a case-by-case basis to identify areas where the problem is particularly severe and where there is significant potential for improvement of system performance.
6. Case Study
In this section, we conduct a case study to demonstrate the feasibility and effectiveness of our approach using the MIMIC-III dataset, a publicly available healthcare dataset containing medical data from over 50,000 hospital visits [9]. We conducted a smaller experiment with 7 classes (ICD codes) and a larger experiment with 40 classes (ICD codes) to emphasize the advantages of the method with different numbers of classes. For our fine-grained classifier, we adopt MedBERT, a variant of BERT trained on EHR data, previously introduced in Section 3.3. Table 2 presents the complexity matrix and architecture parameters of the MedBERT classifier. For both experiments, we split the collected data into 80% training dataset and 20% test dataset. In the training data, we use 5-fold cross-validation to track model performance and select the best performing checkpoints. To avoid overfitting the classifier and take full advantage of MedBERT’s pre-trained knowledge, classifiers were trained with a low learning rate of 5e-5 for 5 epochs, after which the best performing model is selected. In addition, the classifiers were trained using cross-entropy loss. All training and testing processes were performed on a machine with 16 GB of main memory, an Intel Core i7-9700 CPU, and an NVIDIA GeForce RTX 2060 SUPER (8 GB VRAM) GPU.
Table 2: Complexity matrix and parameters of the MedBERT classifier.
| Layer # | Layer Name | Input Size | Output Size |
| 1 | Embedding | 512 | (512, 768) |
| 2 | Encoder 1 self-attention | (512, 768) | (512, 768) |
| 3 | Encoder 1 feed-forward net | (512, 768) | (512, 768) |
| 4-21 | Encoders 2-12 | (512, 768) | (512, 768) |
| 22 | Linear pooling layer | (512, 768) | 768 |
| 23 | Dropout (p = 0.1) | 768 | 768 |
| 24 | Fully connected linear | 768 | # of ICD codes |
| 25 | Softmax activation | # of ICD codes | # of ICD codes |
6.1. Automated Medical Coding Process
Our fine-grained approach performs the medical coding task for one diagnosis at a time. In this experiment, we examine the complete automated medical coding process for predicting the ICD code for a single diagnosis. Suppose the diagnosis to be considered is “Asthma.” The first step in the automated medical coding process is to generate a set of concepts that are semantically related to the diagnosis. In our approach, we generate a list of related concepts by prompting GPT-4 through its Chat Completions API. We send multiple prompts to GPT-4 to guide it in generating categories of related concepts for the diagnosis “Asthma.” Examples of prompts for the diagnosis “Asthma” can be found in Table 1. Figure 10 shows the procedure for promoting GPT-4 and collecting the related concepts for sentence extraction. The prompts listed in Table 1 are sent through the Chat Completions API, which communicates with and receives responses from GPT-4. Each response contains a partial list of related concepts (e.g., symptoms, treatments, etc.). These outputs from GPT-4 are parsed and collected into a complete list of related concepts, which is then passed to the Sentence Extraction module (as shown in Figure 10). The Sentence Extraction module segments the free text in doctor’s notes into sentences and iterates over each sentence. If the current sentence mentions one or more related concepts, the sentence is extracted; otherwise, the sentence is discarded.
After extracting all related sentences from the doctor’s notes, they are combined with the original diagnosis “Asthma” to form a fine-grained data point. Figure 11 shows a portion of the fine-grained data point generated for the “Asthma” diagnosis.

In addition to sentences directly referring to asthma, sentences discussing related terms such as “pulmonary” (lung-related) conditions and “O2” (oxygen) requirements, should also be used to enrich the fine-grained data point, providing additional details that can help with classification. With the generated fine-grained data point, we can now pass it to the hierarchical classifier for ICD code prediction. The hierarchical classifier does not immediately classify the data point, but instead generates the final prediction through a series of subclassifications. Figure 12 shows a hierarchical classification pathway that leads to a medical code for the “Asthma” diagnosis.
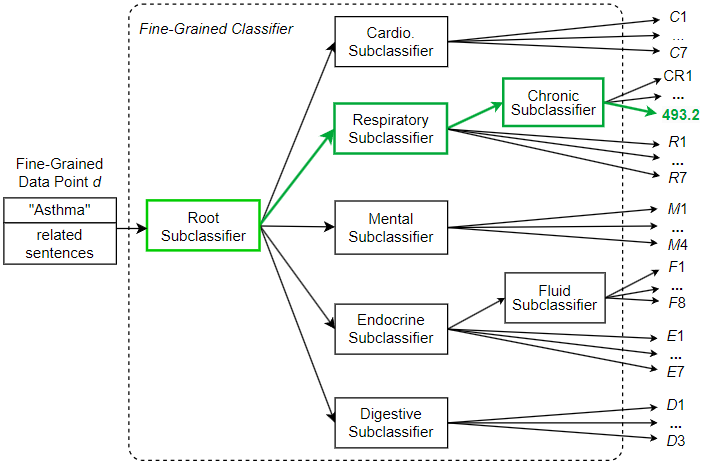
As shown in Figure 12, the fine-grained data point d created for diagnosis “Asthma” is sent to the root subclassifier. This root subclassifier predicts the broad disease category to which the data point belongs. In this example, the root subclassifier recognizes data point d as belonging to the respiratory (breathing-related) disease category and sends d to the Respiratory subclassifier. Then the Respiratory subclassifier further recognizes d as belonging to the chronic disease category and sends d to the Chronic subclassifier. Finally, the leaf Chronic subclassifier recognizes d as ICD code 493.2 (chronic obstructive asthma).
Unlike conventional two-step coarse-grained approaches, the fine-grained approach presented in this section follows several explicit human-understandable steps, including diagnosis selection, sentence extraction, and stepwise hierarchical classification. This information can be provided to the user at any point in the medical coding process to illustrate and justify the method’s medical coding decisions. This additional information maintains a high level of explainability, thereby eliminating the “black box effect” that occurs when a system with a complex architecture focuses solely on deep learning, thus depriving the regular user of important decision-making information.
6.2. Medical Coding with a Small Code Set
In this section, we establish the viability of the approach and examine its performance with a reduced set of medical codes. For this purpose, we employ a set of 7 ICD codes corresponding to closely related heart diseases. In addition, a comparison with a two-step coarse-grained approach was made to demonstrate the improved performance of the proposed method. Although the code set selected contains only a small number of unique codes, the similarities between the various heart diseases poses difficulties for automated ICD coding. In a typical classification task, similar classes are difficult to distinguish, which greatly increases the overall difficulty of classification. Due to class similarities, the classifiers are more likely to confuse the classes, which reduces performance. This is common in ICD coding, as many of the unique codes involved often refer to variants of the same disease. Among the 7 codes examined in this experiment, two of them refer to hypertension, but they do not refer to the same type of hypertension. Code 401.1 refers to benign hypertension, which can be determined by measurement or testing, but without any apparent problematic symptoms. On the other hand, code 401.9 refers to essential hypertension, which can also be determined by measurement and testing, but may be a dangerous condition that requires some form of medical treatment or lifestyle adjustment. Clearly, keeping these two medical codes separable and correctly identifying each is critical to accurate record keeping and effective patient care. Given this particular difficulty, in order to improve coding performance, we opt for a hierarchical classifier design despite the small number of unique codes. That is, we include an additional classification step responsible for separating potentially difficult instances that fall into one of the hypertension classes. Figure 13 shows the hierarchical design of the fine-grained classifier to predict one of the 7 unique ICD codes. As shown in the figure, going through the root subclassifier, a fine-grained data point d may be immediately assigned a final classification and receive the label corresponding to one of the 5 non-hypertension codes in the set. Otherwise, it is assigned to the Hypertension subclassifier in order to differentiate whether the hypertension is benign (code 401.1, “Benign hypertension”) or essential (code 401.9, “Unspecified essential hypertension”). Since the root subclassifier does not need to distinguish the types of suspected hypertension, its classification task becomes simpler and can identify non-hypertension classes more effectively. On the other hand, the Hypertension subclassifier is defined as a dedicated subclassifier trained specifically for separating the two hypertension codes, so it is more capable of predicting one of the two hypertension classes. Under this approach, we split a potentially complex classification task into a series of two less complex subclassification tasks. As a result, each subclassification task has low complexity and high performance, helping to improve the overall classification performance.
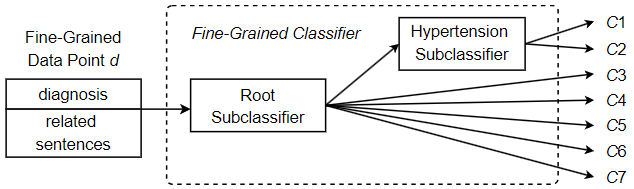
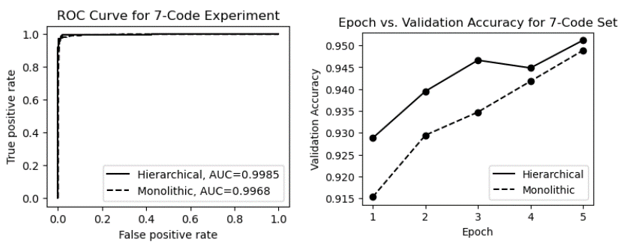
Table 3 shows the performance metrics for processing a dataset with 7 ICD codes using the monolithic and hierarchical approaches. As shown in the table, for this smaller code set, both the monolithic and hierarchical approaches are viable, with an accuracy of over 94.9% and a macro average F1-score of over 87.6%. However, the hierarchical design gives the classifier a slight advantage as it is better able to distinguish between two highly similar hypertension codes.
Table 3: Performance metrics for processing a dataset with 7 ICD codes.
| Method | Accuracy | F1-score | Precision | Recall |
| Monolithic | 0.949 | 0.876 | 0.938 | 0.851 |
| Hierarchical | 0.956 | 0.894 | 0.972 | 0.856 |
The ROC (Receiver Operating Characteristic) curves and epoch vs. validation accuracy plots are presented in Figure 14. The ROC curves in the figure show that the AUC (Area Under the Curve) for both methods is very high, approaching 1.0. As shown in the epoch vs. validation accuracy graphs, both models start out with relatively high performance (thanks to effective pre-training with MedBERT). With fine-tuning, both models showed moderate improvement in accuracy, but neither model demonstrated a significant advantage in validation accuracy.
In addition to these models, we trained and evaluated an approach that employs the conventional two-step or coarse-grained strategy of first vectorizing doctor’s notes documents and then feeding the vectors into a coarse-grained multi-label classifier for ICD code prediction. For comparison, we implemented a coarse-grained multi-labeling approach without a sentence extraction step and fine-grained data point formation to exemplify the advantages offered by these novel components of our proposed approach. We used the same deep learning architecture (MedBERT) to implement this coarse-grained solution with the performance metrics shown in Table 4.
Table 4: Performance metrics for the conventional two-step coarse-grained approach with 7 ICD codes.
| Method | Accuracy | F1-score | Precision | Recall |
| Conventional | 0.539 | 0.460 | 0.375 | 0.644 |
Due to the different nature of multi-label and single-label classification and classification metrics, accurate comparison of specific values is not feasible. However, it is clear from the metrics that there is a significant performance difference between the coarse-grained approach and the two fine-grained approaches. The coarse-grained approach demonstrates more difficult and lower performance classification tasks while the deep learning architecture remains the same. On the other hand, the same deep learning architecture was able to keep up and provide excellent performance when working on the same task in a fine-grained manner. In the next section, we will investigate a larger code set where the performance difference between the two fine-grained approaches (monolithic and hierarchical) is demonstrated.
6.3. Medical Coding with a Large Code Set
Since modern standards such as ICD-9 and ICD-10 contain thousands of unique medical codes, useful automated coding tools must be scalable and able to handle large numbers of codes. In this section, we explore how to apply our fine-grained hierarchical ICD coding approach to a dataset containing 40 unique ICD codes. The expanded code set leads to a more complex label space and a more complex hierarchical classification. Despite the difficulties, an effective automated ICD coding approach should remain robust and usable in terms of classification performance. Our approach employes a hierarchical classification method that divides the complex classifications into multiple subclassifications. Figure 12 shows the design of a hierarchical fine-grained classifier for processing a dataset containing 40 ICD codes. As shown in the figure, predictions are made only after two or three steps of subclassifications. The root subclassifier first accepts the fine-grained data point d and decides which disease family it may belong to. In this experiment, the 40 codes belong to five families, each affecting a different organ system. These five families are cardiovascular, respiratory, endocrine, digestive, and mental. Once the root subclassifier predicts the family of a fine-grained data point, that data point is passed down through the predicted branch. For example, if a data point is classified as an endocrine disease by the root subclassifier, it is passed down to the Endocrine subclassifier for further classification. At this point, some classes can be predicated immediately, resulting in a final ICD code prediction in code set {E1, E2, …, E7}. However, for endocrine diseases associated with body fluids, the data point is sent to the Fluid subclassifier for further classification before an ICD code in code set {F1, F2, …, F8} can be predicted. Table 5 shows the performance metrics for processing a dataset with 40 medical codes using the monolithic and hierarchical approaches. As shown in the table, the hierarchical classification approach remains effective even when the number of unique labels increases significantly. In the case where 40 unique ICD codes need to be predicted, the hierarchical classification method achieved an accuracy and a macro average F1-score of 91.8% and 88.9%, respectively. This successful classification performance provides support for the proposed GPT-enhanced automated coding approach as a potentially useful tool for ICD coding. On the other hand, the performance metrics of the monolithic classifier decreased significantly, with accuracy and macro average F1-score dropping to 73.8% and 70.2%, respectively. This is due to the fact that the subclassifiers of the hierarchical approach have much lower subclassification complexity and correspondingly higher accuracy, whereas the flat or single classifier of the monolithic approach eventually becomes overwhelmed by the increased complexity of the ICD code label space, leading to a degradation of its classification performance.
Table 5: Performance metrics for processing a dataset with 40 ICD codes.
| Method | Accuracy | F1-score | Precision | Recall |
| Monolithic | 0.738 | 0.702 | 0.733 | 0.696 |
| Hierarchical | 0.918 | 0.889 | 0.905 | 0.876 |
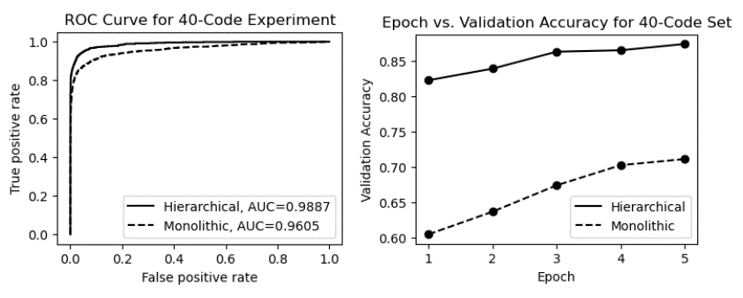
Figure 15 shows the ROC curves and epoch vs. validation accuracy plots for both models. As shown in the figure, the same advantages are demonstrated by the ROC curves and the plots of epoch vs. validation accuracy. Both models gained the base performance from pre-training with MedBERT; however, unlike the previous experiments with a small code set, the hierarchical classifier had a significant performance advantage after the first training epoch (validation accuracy ~0.82 vs. ~0.61). In addition, the ROC curves and AUCs are quite different from previous experiments, which further demonstrates the clear advantage of the hierarchical approach as the number of unique ICD codes (number of classes) increases.
7. Conclusions and Future Work
Despite the many advances in language processing and classification techniques, medical coding remains a challenging task in healthcare. As a classification task, medical coding poses a number of unique challenges that are addressed by the various techniques presented in this paper. Since medical coding is a multi-label classification task, processing doctor’s notes using a fine-grained code assignment method helps limit the label space for individual classifications while still producing an appropriate set of codes. In our approach, we classify only one diagnosis at a time; consequently, individual classifications are simpler and performance is correspondingly higher, even if a large number of unique ICD codes must be considered. Furthermore, we split the prediction process of ICD codes for a given diagnosis into a hierarchical procedure consisting of multiple subclassification steps, thus limiting the complexity of the classification process. Improvements in the hierarchical classifier suggest that classification tasks like ICD coding with related labels (for example, multiple diseases belonging to a shared family) may benefit from having these relations built into the model architecture, as in ICD code hierarchies. To support a fine-grained approach and provide more informative input to the hierarchical classifier used, we have implemented GPT-enhanced sentence extraction, which prompts GPT-4 for related concepts to be used for locating related sentences in doctor’s notes. Unlike previous approaches that use ontologies to generate the related terms used in the sentence extraction step, an approach using GPT-4 provides a useful solution to avoid the additional overhead required to develop a formal medical ontology. The proposed fine-grained hierarchical approach to automated ICD code assignment has yielded promising results in the experiments and provides a basis for effective classification using GPT-enhanced text mining. In addition, the flexibility and effectiveness of GPT-4 in extracting semantically related sentences suggests that there could be further unexplored uses for ICD coding and feature engineering beyond sentence extraction.
While GPT-4 has been used to provide a list of concepts required for our sentence extraction step, the LLM is best known for its wide range of capabilities in a variety of complex tasks. Future work may explore the capabilities of the LLM in automating the various steps of ICD code assignment. For example, GPT-4 could be responsible for the entire sentence extraction step, rather than just generating related concepts. GPT-4 could even be used to perform the entire ICD code prediction, although its performance would need to be carefully examined and characterized. Another potential usage of the generative model is to provide a human-friendly interface for medical coders to answer questions and resolve queries related to predicted codes. Future work could also explore the design and definition of more complex and modular hierarchical classifiers. In particular, improved hierarchical classifiers could employ a variety of decision processes, including decision trees, rule-based reasoning, and deep learning, to produce final classification results. Heterogeneous hierarchies of this type can be designed to be more specific in order to deal with each step of the classification according to the most efficient method. For example, it is usually simple to distinguish between heart disease and eye disease. In most cases, a relatively simple decision-making process can handle this distinction. On the other hand, distinguishing between many highly correlated eye diseases is much more difficult and may require a more powerful decision process, such as the deep learning classifier used in this paper. Since any classification task involving labels can somehow be meaningfully arranged into a hierarchy or taxonomy, future work may explore the application of this hierarchical classification approach not only in ICD coding, but also in different classification tasks such as object recognition, anomaly detection, and topic classification.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgment
We thank the editors and all anonymous referees for spending their valuable time carefully reviewing this paper and making many suggestions for improvement. We also thank the University of Massachusetts Dartmouth for providing financial support to the first author to complete this work.
- WHO, “International statistical classification of diseases and related health problems (ICD),” Health Topics, World Health Organization (WHO), January 1, 2022. Retrieved on December 12, 2023 from: https://www.who.int /standards/classifications/classification-of-diseases
- J. Carberry, H. Xu, “Fine-grained ICD code assignment using ontology-based classification,” In Proceedings of the 2022 IEEE 23rd International Conference on Information Reuse and Integration for Data Science (IRI), 228-233, San Diego, CA, USA, August 2022, doi: 10.1109/ IRI54793.2022.00058.
- I. Goldstein, A. Arzumtsyan, O. Uzuner, “Three approaches to automatic assignment of ICD-9-CM codes to radiology reports,” AMIA Annual Symposium Proceedings, no. 1, 279-283, 2007.
- R. Farkas, G. Szarvas, “Automatic construction of rule-based ICD-9-CM coding systems,” BMC Bioinformatics, 9(suppl 3, S10), April 2008, doi: 10.1186/1471-2105-9-S3-S10.
- J. Medori, C. Fairon, “Machine learning and features selection for semi-automatic ICD-9-CM encoding,” In Proceedings of the NAACL HLT 2nd Louhi Workshop Text Data Mining Health Documents, 84-89, Los Angeles, June 2010.
- M. Li, Z. Fei, F. Wu, Y. Li, Y. Pan, J. Wang, “Automated ICD-9 coding via a deep learning approach,” IEEE/ACM Transactions on Computational Biology and Bioinformatics, 16(4), pp. 1193-1202, July-August 2019, doi: 10.1109/TCBB.2018.2817488.
- T. S. Heo, Y. Yoo, Y. Park, B. Jo, K. Lee, K. Kim, “Medical code prediction from discharge summary: document to sequence BERT using sequence attention,” In Proceedings of the 20th IEEE International Conference on Machine Learning and Applications (ICMLA), 1239-1244, Pasadena, CA, USA, 2021, doi: 10.1109/ICMLA52953.2021.00201.
- J. Carberry, H. Xu, “A hierarchical fine-grained deep learning model for automated medical coding,” In Proceedings of the 3rd International Conference on Computing and Machine Intelligence (ICMI 2024), Central Michigan University, Michigan, USA, April 13-14, 2024.
- A. E. W. Johnson, T. J. Pollard, L. Shen, L. H. Lehman, M. Feng, M. Ghassemi, B. Moody, P. Szolovits, L. Anthony Celi, R. G. Mark, “MIMIC-III, a freely accessible critical care database,” Scientific Data, 3(1), 160035, 2016, doi: doi.org/10.1038/sdata.2016.35.
- P. Chen, S. Wang, W. Laio, L. Kuo, K. Chen, Y. Lin, C. Yang, C. Chiu, S. Chang, F. Lai, “Automatic ICD-10 Coding and Training System: Deep Neural Network Based on Supervised Learning,” JMIR Med Inform, 9(8) :e23230, August 2021, doi: 10.2196/23230.
- Y. Wu, Z. Chen, X. Yao, X. Chen, Z. Zhou, J. Xue, “JAN: Joint attention networks for automatic ICD coding,” IEEE Journal of Biomedical and Health Informatics, 26(10), 5235-5246, October 2022, doi: 10.1109/JBHI.2022.3189404.
- V. Mayya, S. S. Kamath, V. Sugumaran, “LATA – Label attention transformer architectures for ICD-10 coding of unstructured clinical notes,” In Proceedings of the 2021 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), 1-7, Melbourne, Australia, 2021, doi: 10.1109/CIBCB49929.2021.9562815.
- Y. Dong, “A deep learning-driven disease classification method using MIMIC-III database and natural language processing,” In Proceedings of the 2023 IEEE 3rd International Conference on Data Science and Computer Application (ICDSCA), 1068-1072, Dalian, China, October 2023, doi: 10.1109/ICDSCA59871.2023.10392478.
- M. Marin, L. E. Sucar, J. A. Gonzales, R. Diaz, “A hierarchical model for morphological galaxy classification,” In Proceedings of the Twenty-Sixth International Florida Artificial Intelligence Research Society Conference (FLAIRS 2013), 438-443, St. Pete Beach, FL, USA, May 22-24, 2013.
- M. Ramírez-Corona, L. E. Sucar, E. F. Morales, “Hierarchical multi-label classification based on path evaluation,” International Journal of Approximate Reasoning, 68, 179-183, January 2016.
- A. D. Secker, M. N. Davies, A. A. Freitas, J. Timmis, M. Mendao, D. R. Flower, “An experimental comparison of classification algorithms for the hierarchical prediction of protein function,” Expert Update (Magazine of the British Computer Society’s Specialist Group on AI), 9(3), 17-22, 2007.
- K. Wang, S. Zhou, Y. He, “Hierarchical classification of real life documents,” In Proceedings of the 2001 SIAM International Conference on Data Mining (SDM01), 1-16, Chicago, IL, USA, April 5-7, 2001, doi: 10.1137/1.9781611972719.22.
- K. Daisey, S. D. Brown, “Effects of the hierarchy in hierarchical, multi-label classification,” Chemometrics and Intelligent Laboratory Systems, 207, 104177, December 2020, doi: 10.1016/j.chemolab.2020.104177.
- C. N. Silla Jr., A. A. Freitas, “A global-model naive bayes approach to the hierarchical prediction of protein functions,” In Proceedings of the 9th IEEE International Conference on Data Mining, 992-997, December 6-9, 2009, Miami Beach, FL, USA, doi: 10.1109/ICDM.2009.85.
- S. Lawrence, C. L. Giles, A. C. Tsoi, A. D. Back, “Face recognition: a convolutional neural-network approach,” IEEE Transactions on Neural Networks, 8(1), 98-113, January 1997, doi: 10.1109/72.554195.
- X. Peng, T. Tan, T. Fan, “Automatic ICD coding based on bias removal,” In Proceedings of the 2024 IEEE 2nd International Conference on Control, Electronics and Computer Technology (ICCECT), 53-57, Jilin, China, 2024, doi: 10.1109/ICCECT60629.2024.10546138.
- T. Chomutare, A. Budrionis, H. Dalianis, “Combining deep learning and fuzzy logic to predict rare ICD-10 codes from clinical notes,” In Proceedings of the 2022 IEEE International Conference on Digital Health (ICDH), 163-168, Barcelona, Spain, July 2022, doi: 10.1109/ICDH55609.2022.00033.
- A. Kumar, S. Das, S. Roy, “Transfer learning improves unsupervised assignment of ICD codes with clinical notes,” In Proceedings of the 2023 IEEE International Conference on Digital Health (ICDH), 278-287, Chicago, IL, USA, July 2023, doi: 10.1109/ICDH60066.2023.00047.
- N. T. K. Le, N. Hadiprodjo, H. El-Alfy, A. Kerimzhanov, A. Teshebaev, “The recent large language models in NLP,” In Proceedings of the 2023 22nd International Symposium on Communications and Information Technologies (ISCIT), 1-6, Sydney, Australia, October 2023, doi: 10.1109/ISCIT57293.2023.10376050.
- J. Fields, K. Chovanec, P. Madiraju, “A survey of text classification with transformers: how wide? how large? how long? how accurate? how expensive? how safe?” IEEE Access, 12, 6518-6531, 2024, doi: 10.1109/ACCESS.2024.3349952.
- C. Sun, X. Qiu, Y. Xu, X. Huang, “How to fine-tune BERT for text classification?” arXiv:1905.05583 [cs.CL], February 2020, Verson 3, doi: 10.48550/arXiv.1905.05583.
- J. Devlin, M. Chang, K. Lee, K. Toutanova, “BERT: Pre-training of deep bidirectional transformers for language understanding,” arXiv: 1810.04805 [cs.CL], May 2019, Version 2, doi: 10.48550/arXiv.1810.04805.
- F. Karl, A. Scherp, “Transformers are short text classifiers: a study of inductive short text classifiers on benchmarks and real-world datasets,” arXiv:2211.16878v3 [cs.CL], 2023, doi: 10.48550/ arXiv.2211.16878.
- K. M. Yoo, D. Park, J. Kang, S. Lee, W. Park, “GPT3Mix: leveraging large-scale language models for text augmentation,” arXiv:2104.08826 [cs.CL], April 2021, doi: 10.48550/arXiv.2104.08826.
- X. Cai, M. Xiao, Z. Ning, Y. Zhou, “Resolving the imbalance issue in hierarchical disciplinary topic inference via LLM-based data augmentation,” In Proceedings of the 2023 IEEE International Conference on Data Mining (ICDM), 956-961, Shanghai, China, December 2023, doi: 10.1109/ ICDM58522.2023.00107.
- D. W. Otter, J. R. Medina, J. K. Kalita, “A survey of the usages of deep learning for natural language processing,” IEEE Transactions on Neural Networks and Learning Systems, 32(2), 604-624, February 2021, doi: 10.1109/TNNLS.2020.2979670.
- C. Vasantharajan, K. Z. Tun, H. Thi-Nga, S. Jain, T. Rong, C. E. Siong, “MedBERT: a pre-trained language model for biomedical named entity recognition,” In Proceedings of the 2022 Asia-Pacific Signal and Information Processing Association Annual Summit and Conference (APSIPA ASC), 1482-1488, Chiang Mai, Thailand, November 2022, doi: 10.23919/APSIPAASC55919.2022.9980157.
- D. Brin, V. Sorin, A. Vaid, B. S. Glicksberg, A. W. Charney, G. Nadkarni, E. Klang, “Comparing ChatGPT and GPT-4 performance in USMLE soft skill assessments,” Scientific Reports, 13, 16492, 2023, doi: 10.1038/s41598-023-43436-9.
No. of Downloads Per Month
