Brain Tumor Classification Using Deep Neural Network
Volume 5, Issue 5, Page No 765-769, 2020
Author’s Name: Gökalp Çınarer1,a), Bülent Gürsel Emiroğlu2, Recep Sinan Arslan1, Ahmet Haşim Yurttakal1
View Affiliations
1Bozok University, Computer Technologies Department, Yozgat-66100, Turkey
2Kırıkkale University, Computer Engineering Department, Kırıkkale-71451, Turkey
a)Author to whom correspondence should be addressed. E-mail: gokalp.cinarer@bozok.edu.tr
Adv. Sci. Technol. Eng. Syst. J. 5(5), 765-769 (2020); ![]() DOI: 10.25046/aj050593
DOI: 10.25046/aj050593
Keywords: Brain Tumor, Classification, Deep Neural Network
Export Citations
Brain tumors are a type of tumor with a high mortality rate. Since multifocal-looking tumors in the brain can resemble multicentric gliomas or gliomatosis, accurate detection of the tumor is required during the treatment process. The similarity of neurological and radiological findings also complicates the classification of these tumors. Fast and accurate classification is important for brain tumors. Computer aided diagnostic systems and deep neural network architectures can be used in the diagnosis of multicentric gliomas and multiple lesions. In this study, the Deep Neural Network classification model with Synthetic Minority Over-sampling Technique pre-processing was used on the Visually Accessible Rembrandt Images dataset. The proposed model for the classification of brain tumors consists of 1319 trainable parameters and the proposed method has achieved 95.0% accuracy rate. Precision, Recall, F1-measure values are 95.4%, 95.0% and 94.9% respectively. The proposed decision support system can be used to give an idea to doctors in the detection of glioma type tumors.
Received: 13 July 2020, Accepted: 15 September 2020, Published Online: 12 October 2020
1. Introduction
Brain tumors are tumor types located in the brain. The mortality rate is high. There are two different types: benign and malignant. Benign brain tumors, which have a slow growth rate, are easily separable from brain tissue. Malignant brain tumors grow faster and can damage nearby brain tissue[1]. There have been many successful studies in which computer were used in medical applications in recent years. Skin cancer [2], breast cancer [3], thyroid nodules [4] are some of these studies.
In addition, tumors originating from brain tissue are called primary brain tumors, while tumors formed by the splashing of cancerous cells that appear elsewhere in the body are called secondary brain tumors. The most common (65%) and most malignant brain tumor is glioblastoma [5]. Glioblastoma multiforme (GBM) is one of the deadliest cancer types and is a common type of malignant glioma in the central nervous system. With the development of computed tomography and magnetic resonance imaging (MRI), it has become increasingly clear that gliomas may have a multifocal or multicentric [6]. Although the diagnosis of multiple lesions becomes easier with the introduction of computed tomography into the field, the differential diagnosis of multiple cerebral lesions still remains a problem. In multifocal lesions, the lesions are localized in more than one part of the brain and there is a macroscopic or microscopic level relationship between them. In multicentric gliomas, there is neither macroscopic nor microscopic level relationship between lesions. The propagation path of multicentric lesions is unclear, but this situation is slightly different in multiple lesions. Multiple lesions create multiple cerebral areas using a systematic propagation pathway [7]. The correct detection of this condition is very important in the treatment of the disease.
Early diagnosis of brain tumors can change the course of the disease and save lives. On the other hand, computers perform complex operations in a very short time. Computers are used in many areas of life, especially in health, as it produces automatic, fast and accurate results. For this reason, computer-aided diagnostic (CAD) systems are frequently used today. CAD systems analyze the complex relationships in medical data and assist the doctor in decision making [8]. Salvati divided multifocal gliomas into four different categories. These are diffuse, multicentric, multiple and multi-organ. In addition, in his study, he divided his patients into multicentric and multifocal tumor groups and found that there was no significant difference between patients [9]. In a spike case, as malignant glial tumors damage nerve cells, it seems possible that the multicentricity of gliomas may also accompany multiple sclerosis [10]. DNN can learn quickly, automatically identify features, and can be successfully applied in computer learning systems. Besides that, DNNs are also used in some medical image analysis. It can be preferred especially in brain tumor segmentation and classification. It is designed the DNN model to segment brain separate gray and white matter from cerebrospinal fluid and analyzed the segmentation accuracy with DNN [11]. Similarly DNN model for the segmentation of brain tumors is proposed [12].
In this study, Deep Neural Network (DNN) architecture was proposed to classify the MR brain image features as n/a, multifocal, multicentric and gliomatosis. Synthetic Minority Over-sampling Technique (SMOTE) was used as a pre-processing. It has been determined that the pre-processing has a significant effect on the classification success.
2. Material and Methods
2.1. Dataset
In the study, The Repository of Molecular Brain Neoplasia Data (REMBRANDT) dataset [13] was used. The REMBRANDT dataset is a dataset of The Cancer Imaging Archive (TCIA) database shared by the National Cancer Institute. TCIA is an accessible archive containing medical images [14]. This data set contains images of 33 patients. Each patient has an average of 20 MR images, on separate axial, sagittal and coronal planes. For each patient, there are feature key values provided by 3 different neuroradiologists from Thomas Jefferson University (TJU) Hospital. This feature set is called VASARI (Visually Accessible Rembrandt Images). VASARI dataset was used in the classification process.
2.2. SMOTE
SMOTE is a method used in unbalanced data. Every minority class sample is taken and synthetic samples are created by looking at any or the sample’s entire k neighbour. Thus, the minority class is over-sampled [15]. The main difference from other sampling methods is the production of synthetic samples by looking at their neighbours, instead of copying and reproducing the minority class samples. Thus, it realizes class distribution without providing additional information.
2.3. Deep Neural Networks (DNN)
Machine learning technologies potentiate many technologies in the modern world. People use these technologies in many areas such as webcams, e-commerce activities, and mobile devices. In addition, it offers good results in complex problems. These are identifying objects from images, converting speech to text and matching web contents with the interests of the users. Traditional ML techniques have some limitations in processing raw data. Extracting features representing data in the raw form and converting them into feature vectors used as input for classifier is a difficult process that requires expertise [16].
Deep learning allows abstract representation of data by creating simple but nonlinear modules. In this way, it is possible to learn very complex functions. This structure reveals the distinguishing features between classes for classification problems. Deep learning produces successful results for many problems that cause problems in the advancement of artificial intelligence methods. It is used in many areas such as image recognition [17-19], speech processing [20, 21], and gene mutation prediction [22]. It is thought that DNN will make faster progress thanks to the new neural network structures proposed with an increase in computation capacity and amount of data [16].
Deep neural network structure is shown in Figure 1.
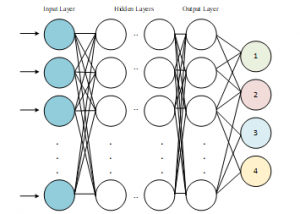
Figure 1: Deep Neural Network Schema [16]
The development of computer hardware structure and the widespread use of GPU architecture facilitates and speed up the training of the complex and multi-layered neural networks as shown in Figure 1. In this way, more outstanding [23].
The mathematical model used in the neural network training stage is shown in Eq-1.
![]()
According to Equation, represents the weight matrix in the nth layer and is activation function. This structure gives better results than single-layer networks [24].
In this study, a deep neural network design with 1 input, 1 output and 2 hidden layer was designed.
The increasing of software technologies shaped under three specialized topics such as artificial intelligence, machine learning and deep learning has caused an increase in interest in these three subjects. Deep Neural Networks finds the relationship between a range of inputs and outputs. Successful results in many areas such as image processing, natural language processing, voice recognition [25].
3. Results
The application was carried out on the open source Python program. Tensorflow and Keras libraries were used. The count plot of the data to be classified into classes is given in Figure 2.
All images included in the Rembrandt data set consist of axial, sagittal and coronal image planes with an average of 20 images per plane for a total of 33 patients. In the data set, 30 different features were obtained from each tumor brain MR image. Rembrandt images (VASARI) were analyzed by three TJU radiologists and features of the images were extracted. In this way, a data set
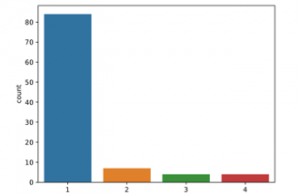
Figure 2: Count plot of data before SMOTE

Figure 3: Count plot of data after SMOTE
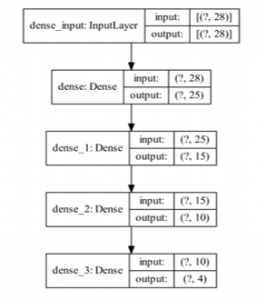
Figure 4: Proposed DNN Model
consisting of 99 patients and 30 different characteristic features used in the study. The data were classified according to n/a, multifocal, multicentric and gliomatosis. The data appears to be unbalanced. (84 n/a patient, 7 multifocal patient, 4 multicentric patient and 4 gliomatosis patient). For this reason, SMOTE pre-treatment was applied. Figure 3 shows the count plot after the SMOTE pre-processing.
The proposed DNN model is given in Figure 4. There are 3 hidden layers in the model, having 25, 15, and 10 filters, respectively. Model includes 1319 trainable parameters.
The activation function used in hidden layers is ReLu. The activation function used in the output layer is SoftMax. The optimizer who used the error back propagation phase is Adam. In this study, the data set was formed as 70% training and 30% test group. Figure 5 shows the change in accuracy on training and test data over 100 epochs.
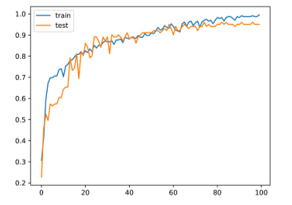
Figure 5: Accuracy graph
As can be seen from the graphic, the proposed method has overcome the over-fitting problem. Figure 6 shows confusion matrix.
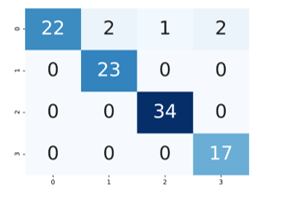
Figure 6: Confusion matrix
DNN model was used to categorize brain tumors into four classes (n / a, multifocal, multicentric and gliomatosis). Four basic measurement indicators were used when evaluating the classification results. The proposed DNN classifier knew the opinion of radiologists with 95% accuracy. The success rate of the model was also examined with other performance metrics such as precision recall and F1. Respectively Precision, Recall, F1-measure values are 0.954, 0.950 and 0.949.
4. Discussion
Deep learning methods are used in many research fields of medical image analysis, from image recording, radiomic feature selection, segmentation and classification. Estimating the type of brain tumors is quite difficult and troublesome in the medical field.The aim of this study is to classify MR images of brain tumors as n/a, multifocal, multicentric and gliomatosis. In this study, unlike other studies, multiple cerebral lesions were classified. These lesions can have similar features and are difficult to classify. Studies in the literature determine tumor levels or predict whether the tumor is benign or malignant. Kumar and Vijay Kumar classified brain images as tumors and non-tumors using the Feed Forwarded Artificial neural network-based classifier and reached 91.17% accuracy with the ensemble classifier [26]. Glioblastoma and low-grade gliomas using logistic regression algorithm is classified with 0.88 accuracy [27]. Mahajani classified the MRI brain image as normal or abnormal with Back Propagation Neural Network (BPNN) and K Nearest Neighbor (KNN) algorithms. 70% accuracy using the KNN classifier and 72.5% accuracy using the BPNN classifier [28]. In the literature this dataset was tested with KNN, Random Forest (RF), Support Vector Machines (SVM), Linear Discriminant Analysis (LDA) machine learning algorithms. SVM algorithm with 90% accuracy rate was found to be better compared to other algorithms [29]. As seen in the examples, machine learning algorithms are commonly used approaches in classification. However, since deep learning models can analyze complex relationships effectively, it has gained an important place in machine learning in recent years. In another study, VGG-19 architecture used to classify brain tumors grades (I, II, III and IV) and achieve 0.90 accuracy and 0.96 average precision [30]. Also, in the other study it is classified the grades of glioma tumor using Convolutional Neural Networks (CNN) with an accuracy of 91.16% [31]. Compared to other algorithms, it has been seen that the applied method has advantages over traditional classifier algorithms. The complexity of the brain structure in the analysis of the brain image and the lack of imaging standards prevent the correct application of computer-aided learning systems in classifying brain tumors. Therefore, radiologist views come to the fore in determining these criteria. Table 1 shows some deep learning studies in the literature.
Table 1: Deep learning approaches for brain tumor classification
| Aim and Method | Accuracy |
| Tumor Segmentation-DNN[32] | 0.88 |
| Brain Tumour, CNN[33] | 91.43 |
| Brain Tumour, Capsule Network[34] | 86.56 |
| Brain Tumoru, DNN[35] | 90.66 |
In addition, proposed classification model can be tested with different parameters and radiomic features. Obtaining the data by experienced radiologists and determining the radiologist views with high accuracy is an indicator of the success of the algorithm. The Deep Learning approach facilitates researchers and trains the model using its own hyper parameters without specifying many features beforehand. But it is very important to apply the correct parameters to the model. In this study, the deep learning model was used and high accuracy was obtained with the correct parameters applied.
5. Conclusion
In the article, multifocal, multicentric and gliomatosis classification of brain tumors with SMOTE pre-processing of the DNN model were examined. The results show that the proposed method is very successful. The proposed method gives automatic, fast and accurate results. It can give an idea as a second look when doctors are unstable.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgment
No funding,
- J.C. Buckner, P.D. Brown, B.P. O’Neill, F.B. Meyer, C.J. Wetmore, J.H. Uhm, “Central nervous system tumors,” in Mayo Clinic Proceedings, 2007, doi:10.4065/82.10.1271.
- N. Razmjooy, M. Ashourian, M. Karimifard, V. V. Estrela, H.J. Loschi, D. do Nascimento, R.P. França, M. Vishnevski, “Computer-Aided Diagnosis of Skin Cancer: A Review,” Current Medical Imaging Formerly Current Medical Imaging Reviews, 2020, doi:10.2174/1573405616666200129095242.
- J.Y. Kim, J.J. Kim, L. Hwangbo, H.B. Suh, S. Kim, K.S. Choo, K.J. Nam, T. Kang, “Kinetic heterogeneity of breast cancer determined using computer-aided diagnosis of preoperative MRI scans: Relationship to distant metastasis-free survival,” Radiology, 2020, doi:10.1148/radiol.2020192039.
- D. Fresilli, G. Grani, M.L. De Pascali, G. Alagna, E. Tassone, V. Ramundo, V. Ascoli, D. Bosco, M. Biffoni, M. Bononi, V. D’Andrea, F. Frattaroli, L. Giacomelli, Y. Solskaya, G. Polti, P. Pacini, O. Guiban, R. Gallo Curcio, M. Caratozzolo, V. Cantisani, “Computer-aided diagnostic system for thyroid nodule sonographic evaluation outperforms the specificity of less experienced examiners,” Journal of Ultrasound, 2020, doi:10.1007/s40477-020-00453-y.
- H. Ohgaki, P. Kleihues, Epidemiology and etiology of gliomas, Acta Neuropathologica, 2005, doi:10.1007/s00401-005-0991-y.
- C.G. Patil, P. Eboli, J. Hu, Management of Multifocal and Multicentric Gliomas, Neurosurgery Clinics of North America, 2012, doi:10.1016/j.nec.2012.01.012.
- R.P. Thomas, L.W. Xu, R.M. Lober, G. Li, S. Nagpal, “The incidence and significance of multiple lesions in glioblastoma,” Journal of Neuro-Oncology, 2013, doi:10.1007/s11060-012-1030-1.
- T.R. Jensen, K.M. Schmainda, “Computer-aided detection of brain tumor invasion using multiparametric MRI,” Journal of Magnetic Resonance Imaging, 2009, doi:10.1002/jmri.21878.
- M. Salvati, L. Cervoni, P. Celli, R. Caruso, F.M. Gagliardi, “Multicentric and multifocal primary cerebral tumours. Methods of diagnosis and treatment,” Italian Journal of Neurological Sciences, 1997, doi:10.1007/BF02106225.
- L.C. Werneck, R.H. Scola, W.O. Arruda, L.F.B. Torres, “Glioma and multiple sclerosis: case report.,” Arquivos de Neuro-Psiquiatria, 2002, doi:10.1590/S0004-282X2002000300024.
- P. Moeskops, M.A. Viergever, A.M. Mendrik, L.S. De Vries, M.J.N.L. Benders, I. Isgum, “Automatic Segmentation of MR Brain Images with a Convolutional Neural Network,” IEEE Transactions on Medical Imaging, 2016, doi:10.1109/TMI.2016.2548501.
- P. Dvořák, B. Menze, “Local structure prediction with convolutional neural networks for multimodal brain tumor segmentation,” in Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), 2016, doi:10.1007/978-3-319-42016-5_6.
- L. Scarpace, A.E. Flanders, R. Jain, T. Mikkelsen, D.W. Andrews, “Data From REMBRANDT,” The Cancer Imaging Archive, 2015, doi:10.7937/K9/TCIA.2015.588OZUZB.
- K. Clark, B. Vendt, K. Smith, J. Freymann, J. Kirby, P. Koppel, S. Moore, S. Phillips, D. Maffitt, M. Pringle, L. Tarbox, F. Prior, “The cancer imaging archive (TCIA): Maintaining and operating a public information repository,” Journal of Digital Imaging, 2013, doi:10.1007/s10278-013-9622-7.
- N. V. Chawla, K.W. Bowyer, L.O. Hall, W.P. Kegelmeyer, “SMOTE: Synthetic minority over-sampling technique,” Journal of Artificial Intelligence Research, 2002, doi:10.1613/jair.953.
- W. Liu, Z. Wang, X. Liu, N. Zeng, Y. Liu, F.E. Alsaadi, “A survey of deep neural network architectures and their applications,” Neurocomputing, 2017, doi:10.1016/j.neucom.2016.12.038.
- H. Fujiyoshi, T. Hirakawa, T. Yamashita, Deep learning-based image recognition for autonomous driving, IATSS Research, 2019, doi:10.1016/j.iatssr.2019.11.008.
- G. Hu, C. Yin, M. Wan, Y. Zhang, Y. Fang, “Recognition of diseased Pinus trees in UAV images using deep learning and AdaBoost classifier,” Biosystems Engineering, 2020, doi:10.1016/j.biosystemseng.2020.03.021.
- M. Trokielewicz, A. Czajka, P. Maciejewicz, “Post-mortem iris recognition with deep-learning-based image segmentation,” Image and Vision Computing, 2020, doi:10.1016/j.imavis.2019.103866.
- A.L. Maas, P. Qi, Z. Xie, A.Y. Hannun, C.T. Lengerich, D. Jurafsky, A.Y. Ng, “Building DNN acoustic models for large vocabulary speech recognition,” Computer Speech and Language, 2017, doi:10.1016/j.csl.2016.06.007.
- J. Novoa, J. Fredes, V. Poblete, N.B. Yoma, “Uncertainty weighting and propagation in DNN–HMM-based speech recognition,” Computer Speech and Language, 2018, doi:10.1016/j.csl.2017.06.005.
- M.K.K. Leung, H.Y. Xiong, L.J. Lee, B.J. Frey, “Deep learning of the tissue-regulated splicing code,” Bioinformatics, 2014, doi:10.1093/bioinformatics/btu277.
- A. Yiğit, “İŞ SÜREÇLERİNDE İNSAN GÖRÜSÜNÜ DERİN ÖĞRENMEİLE DESTEKLEME,” Trakya Üniversitesi, Fen Bilimleri Enstitüsü, Yayınlanmış Yüksek Lisans Tezi, 2017.
- G.E. Dahl, T.N. Sainath, G.E. Hinton, “Improving deep neural networks for LVCSR using rectified linear units and dropout,” in ICASSP, IEEE International Conference on Acoustics, Speech and Signal Processing – Proceedings, 2013, doi:10.1109/ICASSP.2013.6639346.
- Y. Zhang, M. Pezeshki, P. Brakel, S. Zhang, C. Laurent, Y. Bengio, A. Courville, “Towards end-to-end speech recognition with deep convolutional neural networks,” in Proceedings of the Annual Conference of the International Speech Communication Association, INTERSPEECH, 2016, doi:10.21437/Interspeech.2016-1446.
- P. Kumar, B. Vijay Kumar, “Brain tumor MRI segmentation and classification using ensemble classifier,” International Journal of Recent Technology and Engineering, 2019.
- K.L.C. Hsieh, C.M. Lo, C.J. Hsiao, “Computer-aided grading of gliomas based on local and global MRI features,” Computer Methods and Programs in Biomedicine, 2017, doi:10.1016/j.cmpb.2016.10.021.
- P.P.S. Mahajani, “Detection and Classification of Brain Tumor in MRI Images,” International Journal of Emerging Trends in Electrical and Electronics, 2013.
- G. Cinarer, B.G. Emiroglu, “Classificatin of Brain Tumors by Machine Learning Algorithms,” in 3rd International Symposium on Multidisciplinary Studies and Innovative Technologies, ISMSIT 2019 – Proceedings, 2019, doi:10.1109/ISMSIT.2019.8932878.
- M. Sajjad, S. Khan, K. Muhammad, W. Wu, A. Ullah, S.W. Baik, “Multi-grade brain tumor classification using deep CNN with extensive data augmentation,” Journal of Computational Science, 2019, doi:10.1016/j.jocs.2018.12.003.
- S. Khawaldeh, U. Pervaiz, A. Rafiq, R.S. Alkhawaldeh, “Noninvasive grading of glioma tumor using magnetic resonance imaging with convolutional neural networks,” Applied Sciences (Switzerland), 2017, doi:10.3390/app8010027.
- M. Havaei, A. Davy, D. Warde-Farley, A. Biard, A. Courville, Y. Bengio, C. Pal, P.M. Jodoin, H. Larochelle, “Brain tumor segmentation with Deep Neural Networks,” Medical Image Analysis, 2017, doi:10.1016/j.media.2016.05.004.
- J.S. Paul, A.J. Plassard, B.A. Landman, D. Fabbri, “Deep learning for brain tumor classification,” in Medical Imaging 2017: Biomedical Applications in Molecular, Structural, and Functional Imaging, 2017, doi:10.1117/12.2254195.
- P. Afshar, A. Mohammadi, K.N. Plataniotis, “Brain Tumor Type Classification via Capsule Networks,” in Proceedings – International Conference on Image Processing, ICIP, 2018, doi:10.1109/ICIP.2018.8451379.
- D. Nie, H. Zhang, E. Adeli, L. Liu, D. Shen, “3D deep learning for multi-modal imaging-guided survival time prediction of brain tumor patients,” in Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), 2016, doi:10.1007/978-3-319-46723-8_25.
