Dissection of Quantitative Trait Loci (QTL), annotation of Single Nucleotide Polymorphism (SNP), and Identification of Candidate Genes for Grain Yield in Triticum turgidum L. var durum
Dissection of Quantitative Trait Loci (QTL), annotation of Single Nucleotide Polymorphism (SNP), and Identification of Candidate Genes for Grain Yield in Triticum turgidum L. var durum
Volume 5, Issue 5, Page No 1020-1027, 2020
Author’s Name: Issame Farouk1,a), Ahmad Alsaleh2, Jihan3, Fatima Gaboun4, Bouchra Belkadi1, Abdelkarim Filali Maltouf1, Zakaria Kehel3, Ismahane Elouafi5, Nasserelhaq Nsarellah4, Dimah Habash6, M. Miloudi Nachit3
View Affiliations
1Laboratory of Microbiology and Molecular Biology, Department of Biology, Faculty of Sciences, Med V University, Rabat, 10000, Morocco
2Dept. Science and Technology Bozok University, Yozgat, 66100, Turkey
3ICARDA. Av. Med Belarbi Alaoui, BP6299, Al Irfane, Rabat, 10000, Morocco.
4INRA. Institut National de Recherche Agronomique Av. Med Belarbi Alaoui, Al Irfane, Rabat, 10000, Morocco
5ICBA. Dubai, 14660, United Arab Emirates
6Securewheat, AL3, St. Albans, UK
a)Author to whom correspondence should be addressed. E-mail: farouk@gmail.com
Adv. Sci. Technol. Eng. Syst. J. 5(5), 1020-1027 (2020); ![]() DOI: 10.25046/aj0505125
DOI: 10.25046/aj0505125
Keywords: Durum wheat, SNP, Candidate genes, Grain yield, QTL, Linkage map
Export Citations
Durum wheat (Triticum turgidum L. var durum) is among the most important crops in the world. High and stable grain yield in diverse environments is the major objective in durum breeding programs. This trait is linked to the quantitative trait loci (QTL). For the detection of QTL linked to the grain yield, it is necessary to construct a high-density genetic linkage map. The aims of this study were to detect the candidate genes comprised in the QTLs on 2A chromosome linked to grain yield and annotate the single nucleotide polymorphisms (SNPs). The linkage map of Lahn/Cham1 population was used to identify QTLs. In multi-environmental analysis and employing bioinformatic approaches, 583 sequences corresponding to the SNPs markers selected from the detected QTL regions were analyzed, 122 SNP sequences were annotated of which 53% of the candidate genes were involved in stresses tolerance, 29.5% in plant development and growth, and 3.3% in cell transport. Moreover, 1.6% of candidate genes were retrotransposon and transposon 2.4% with unknown function. Further 9.8% were related in other cellular processes. The results also showed that 66.7% of the candidate genes harbored on 4B chromosome, were involved in stresses tolerance and 33.3% in plant development and growth. Additionally, in the specific and stressed environments analysis, the DNA sequences of the four QTL detected on 2A chromosome were used for homology search, 546 candidate genes were identified of which some were present in several QTL (F-box gene family, hydroxyproline-rich glycoprotein-like), retrotransposons and transposons and others. This study provided information on employing SNPs markers to detect candidate genes linked with grain yield trait in durum wheat in contrasting environments (dry, cold, hot).
Received: 29 June 2020, Accepted: 04 October 2020, Published Online: 20 October 2020
1. Introduction
Durum wheat (Triticum turgidum L. var durum) is a tetraploid wheat, its genome is constituted by seven homoeologous groups of chromosomes A and B. Its grain use is reported in [1, 2]. The nutritional values for durum grain were published in earlier research works [1-3]. Grain yield is related to the QTL additive effects and to their interactions with the environment. Several wheat genetic linkage maps were constructed with various molecular markers, such as Restriction Fragment Length Polymorphism (RFLP), Amplified Fragment Length Polymorphism (AFLP), Simple Sequence Repeats (SSR) and other markers [4-13]. The construction of saturated maps with large number of molecular markers of Single Nucleotide Polymorphism
|
aURGI : Unité de Recherche Génomique info bQTL : Quantitative trait loci cNCBI: National Center for Biotechnology Information, the USA dBlast: Basic Local Alignment Search Tool |
| 2A DNA sequence downloaded from URGIa |
| QTLb sequence isolation (using the flanked markers and GeneStudio software) |
| NCBIc blastd against Rice and Brachypodium genomes |
| Candidate gene identification |
(SNP) [14-16] and Diversity Array Technology (DArT) [10, 17-23] are fundamental to detect candidate genes for desirable traits in crops, such as durum wheat. Both markers (SNP and DArT) are small sequence of DNA. These sequences can be compared with the public databases [URGI (Unité de Recherche Génomique Info) Blast of the Institut National de la Recherche Agronomique (INRA, France) and NCBI (National Center for Biotechnology Information, the USA)] to identify the candidate genes involved in a QTL controlling a specific trait. Recently the markers (SNP and DArT) were used for QTLs regions detection and for the identification of the candidate genes involved in these QTLs of wheat [24-29]. The high and stable grain yield across diverse environments constitutes the principal aim for durum breeding programs.
The aims of this work were to: 1) identify the SNPs mapped into QTL regions; 2) extend the SNPs sequence analysis in terms of functional and comparative analysis; and 3) identify the candidate genes constituting the QTLs harbored by 2A chromosome and linked to grain yield.
2. Materials and methods
2.1. Plant materials
|
a SNP : Single Nucleotide polymorphism bURGI : Unité de Recherche Génomique info c Blast: Basic Local Alignment Search Tool d NCBI: National Center for Biotechnology Information, the USA Similarity presence in URGI No similarity presence in URGI
|
| SNPa sequence |
| URGIb blastc |
| Sequence expansion |
| NCBId blastc |
| Candidate gene identification |
The genetic population used in this study (Lahn/Cham1) was published in [8, 30] and presented in the conference [31]. The population was phenotyped for grain yield in 11 contrasting environments (cold / drought / heat). The characteristics of the parents: Lahn and Cham1 were described in earlier paper [8, 30].
The Lahn/Cham1 genetic map that consisted of 1425 SNPs, 216 SSR and 31 other molecular markers was constructed using IciMapping program v 4.1.0.0 was presented in [31].
The QTLs linked to grain yield detected using this map were used for candidate genes identification and SNPs annotation.
2.2. SNPs annotation and candidate genes identification
The methodology used for the SNPs annotation and the candidate genes identification is as flowing:
For the QTLs detection in specific and stressed environments, 9 QTLs were detected, 4 of which were localized on 2A, due to its strong contribution to grain yield [32, 33], the full DNA sequence of 2A chromosome were downloaded from URGI and the 4 DNA sequences corresponding to the 4 QTL were isolated based on markers flanking of these QTL regions [24, 25, 29] using the GeneStudio software. More informations about the environments, DNA sequences size, environmental conditions, QTL intervals and flanking markers are summarized in Table 1. Thereafter, the sequences isolated were served as queries for Blast search in NCBI against Rice and Brachypodium genomes to identify the candidate genes composing these QTLs detected in the four environments (EP, INC, RF, and RQ), see Figure 1
In multi- environmental analysis, the 583 SNPs selected from the QTLs regions and harbored by durum chromosomes [31] were analyzed through Blast (Basic Local Alignment Search Tool) search homology using the URGI Blast of (INRA), France (https://urgi.versailles.inra.fr/blast/blast.php) with the aim to expand the SNP sequences in terms of length [34] (Figure 2).
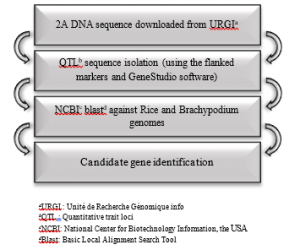
Figure 1: Candidate genes identification in specific and stressed environments
Subsequently, the derived genomic sequences (280bp) were served as query to identify the candidate genes using Blast search homology in NCBI (National Center for Biotechnology Information, the USA), see Figure 2. The Venn diagram was used to present the results.
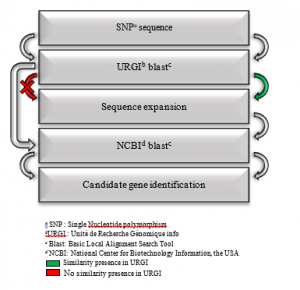
Figure 2: Candidate genes identification in multi-environmental analysis
More details about the candidate genes identified in combined and in specific environments QTLs analysis are shown in supplementary materials S1 and S2, respectively.
Table 1: QTL detected on 2A chromosome with the environmental conditions, QTL peak positions in cM, QTL regions interval size in centimorgan (cM) and in Mega bases (Mb) and with QTLs region flanking markers.
| Environment | Environmental Conditions | Peak position (cM) |
QTLa interval size |
Flanking markers | ||
| cM | Mb | Left marker | Right marker | |||
| EPb | Cold | 142 | 5.02 | 1.33 | wsnpf_2280363 | wmcg177bp190 |
| INCc | Drought (vetch incorporation) | 046 | 5.90 | 2.64 | wsnpf_998898 | cnlh127bp435 |
| RFd | Drought | 270 | 3.59 | 4.57 | wsnpf_1000185 | gwmi312bp189 |
| RQe | Heat | 063 | 6.49 | 0.10 | wsnpf_2276833 | gwmi636bp111 |
aQTL: Quantitative Trait Loci
bEP: Early planting at Tel Hadya / Syria (ICARDA main station), 36°0.1’N 36°56’E, 284m, the sowing is in mid-October, 330mm of rainfall and 70mm of irrigation at sowing, the temperatures were 2 °C at Tillering and 20 °C at flowering
cINC: Incorporated in field (vetch biomass incorporation in soil) at same station, the sowing date in Mid-November with 330mm as rainfall, the temperatures were 8 oC at tillering and 24 oC at flowering
dRF: Rainfed (vetch above biomass harvested, no incorporation in soil) at the same station, sowing in Mid-November, the rainfall temperatures are also the same
eRQ: Raqqa station (35°57’ N 39°0’E, 295m) with chickpea rotation, sowing in Mid-November, the rainfall was 150mm and 450 mm of irrigation, were 10 oC at tillering and 30 oC at flowering
fSNP: Single Nucleotide Polymorphism
gwmc: Wheat Microsatellite Consortium (SSR)
hcnl: Cornell University (EST-SSR)
igwm: Gatersleben Wheat Microsatellite (SSR)
- Results
3.1. Candidate genes detection in specific and stressed environments
The search for candidate genes in the QTLs corresponding to the environments (dry / cold / heat) show the presence of 407 candidate genes in Rice as in Brachypodium databases. Moreover, in the Rice database 139 genes identified were retrotransposons and transposons that have showed no correspondence in Brachypodium database.
3.1.1. Dry environment
As for the dry environments (RF and INC), the QTLs detected in these environments (Table 2) spanned on 4.57 Mb for RF and 2.64 Mb for INC. In RF (dryland) were identified 147 candidate genes and in INC (dryland with incorporation of vetch crop) 376 candidate genes, including genes involved in stress tolerance (biotic and abiotic stress) and in other cellular processes, the retrotransposons and transposons were also present in the two QTLs with a high frequency especially in the RF environment (Supplementary: Table S1). Also, it was noticed that the incorporation of legume vetch in the soil increased the expression of candidate genes. This expression is higher in dryland with vetch incorporation in soil (INC) than in dryland without vetch incorporation in the soil (RF). This may be due to increased availability of nitrogen and water through the organic matter incorporation in the soil.
3.1.2. Cold environments
In the cold environment (EP) characterized by low temperature during the tillering developmental stage (Table 2), the QTL detected was spanned on 1.33 Mb, it contained 17 candidate genes among those involved in plant development and abiotic/biotic stress tolerance: as Germin-like protein 8-14, protein far1-related sequence 5, putative FBD-associated F-box protein, Photosystem II 44 kDa protein and others. The retrotransposons and transposons are also present in this QTL (Supplementary: Table S1).
Table 2: The environmental conditions, the number of candidate genes identified for the four QTL linked to grain yield of the four stressed environments (EP, INC, RF and RQ) with the QTL interval size, and the number of candidate genes identified for Rice and Brachypodium
| Environment | Environmental Conditions | QTLa Interval size (Mb) | Candidates genes number | |
| Rice | Brachypodium | |||
| EPb | Cold | 1.33 | 017 | 012 |
| INCc | Drought (vetch incorporation) | 2.64 | 147 | 113 |
| RFd | Drought | 4.57 | 376 | 277 |
| RQe | Heat | 0.10 | 006 | 005 |
aQTL: Quantitative Trait Loci
bEP: Early planting at Tel Hadya / Syria (ICARDA main station), 36°0.1’N 36°56’E, 284m, the sowing is in mid-October, 330mm of rainfall and 70mm of irrigation at sowing, the temperatures were 2 °C at Tillering and 20 °C at flowering
cINC: Incorporated in field (vetch biomass incorporation in soil) at same station, the sowing date in Mid-November with 330mm as rainfall, the temperatures were 8 oC at tillering and 24 oC at flowering
dRF: Rainfed (vetch above biomass harvested, no incorporation in soil) at the same station, sowing in Mid-November, the rainfall temperatures are also the same
eRQ: Raqqa station (35°57’ N 39°0’E, 295m) with chickpea rotation, sowing in Mid-November, the rainfall was 150mm and 450 mm of irrigation, were 10 oC at tillering and 30 oC at flowering
Table 3: SNPs annotated and not annotated in A and B genomes
| Genome | SNPs | |||
| Annotated | Not annotated | Total | Percentage of SNPs annotated (%) | |
|
1A 2A 3A 4A 5A 6A 7A |
15 17 6 8 1 5 7 |
24 63 33 31 11 14 46 |
39 80 39 39 12 19 53 |
38.46 21.25 15.38 20.51 08.33 26.31 13.20 |
| A genome | 59 | 222 | 281 | 20.99 |
|
1B 2B 3B 4B 5B 6B 7B |
17 16 2 9 5 13 1 |
35 63 10 64 28 29 10 |
52 79 12 73 33 42 11 |
32.69 20.25 16.66 12.32 15.15 30.95 09.10 |
| B genome | 63 | 239 | 302 | 20.86 |
3.1.3. Hot environments
Further, the QTL detected in the hot environment (RQ) which is characterized by high temperatures during the anthesis developmental stage compared with the other environments (Table 2), the QTL region was spanned on a small interval of 0.1Mb that contained 6 genes, the first one is an LRR receptor-like serine/threonine-protein kinase gene involved in plant resistance to pathogens and the others are a retrotransposons and transposons (Supplementary: Table S1).
3.2. Mapping of SNPs to identify candidate genes
Of the mapped SNPs, 80 were mapped on 2A. Additional information for other chromosomes is shown in Table 3.
As for the candidate genes number, the 2A along 1B harbored the highest number (17), while in terms of annotation percentage for other chromosomes see Table 3.
The SNPs sequences availability provides the possibility of identifying gene loci contributing to grain yield.
Among the 583 SNP sequences selected, 122 sequences were annotated. the candidate genes involved in various growth, development processes, and stress tolerances have been identified, of which 77 sequences were enlarged with URGI database and annotated with NCBI. Whereas the remaining 45 SNPs sequences were annotated directly with NCBI, as no-similarity presence was available to enlarge in URGI database (Figure 3). Several sequences did not show any similarity in URGI and NCBI databases
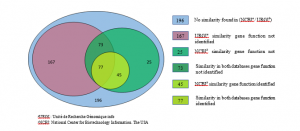
Figure 3: Annotation and similarity study on Venn diagram of SNP.
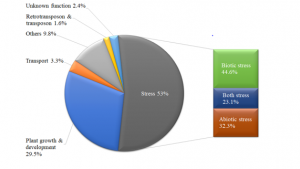
Figure 4: Functional classification of candidate genes co-located with QTLs linked to grain yield.
The 583 SNPs marked on Venn diagram (Figure 3) showed that 77 sequences were annotated (candidate genes identified) with similarity presence with both databases; while 45 sequences were annotated with similarity presence only in NCBI. Further, 73 were not annotated but had similarity in both databases. Also 167 SNP had similarity only in URGI and 25 only in NCBI, consequently both sets of SNPs were not annotated). The remaining196 sequences were not annotated, showing no similarity in the two databases.
The analysis with bioinformatic tools showed that 53% of candidate genes found were related to stresses tolerance, with 44.6% of them to biotic stress [powdery mildew, stem rust (sr32), leaf rust (lr10, lr34)], 32.3% to abiotic stress (cold heat, drought, salinity) and 23.1% to both stresses; 29.5% to plant development and growth; 3.3% to cell transport, and 1.6% to retrotransposon and transposon; and only 2.4% were with unknown function.
Further, 9.8% were involved in other cellular processes (secondary metabolism, pigments, herbicide detoxification, wall formation, etc.), see Figure 4. More informations about the SNPs annotation and candidate gene functions were shown in the supplementary table (S2).
Some candidate genes are involved in several processes, such as the basic helix-loop-helix (bHLH) proteins pertaining to the superfamily of transcription factors (TFs) and linked with wsnp_3026224. These proteins have been linked with the regulation of diverse biological processes as growth, development, and response to diverse stresses.
Moreover, in the 4BS, 57.1% of candidate genes found, were linked to stresses tolerance and 42.9% in plant development and growth.
4. Discussion
4.1. Candidate genes detection in specific and stressed environments
We found among 9 QTLs related to grain yield, four were located on 2A chromosome, these QTLs were detected in diverse stressed environments: EP characterized by cold, INC and RF by drought, and RQ by heat. These results are in accordance with several studies on grain yield QTLs located on 2A chromosome [24, 32, 33].
4.1.1. Dry environment
In the dry environment (INC), among the candidate genes were serpin-Z5, Probable glucuronosyltransferase Os04g0103100, Chalcone synthase 1, Fatty acid amide hydrolase isoform X1, Amino acid permease 3 (Supplementary: Table S1). This is in the agreement with the study of [30] where the transcriptomic pattern under progressive water stress of the two parents: Lahn and Cham1 (parents of the current studied population) and one recombined inbred line where the candidate genes cited above showed an overexpression. Such as the candidate gene coding for the mitochondrial metalloendopeptidase OMA1 identified in the QTL of the dry environment RF (Supplementary: Table S1). This was also confirmed by [30]. Similarly, the F-box and associated protein family were overexpressed in the dry environments RF and INC (Supplementary Table S1), but under cold stress was downexpressed [35]. Some genes of this large family under abiotic stress may be over or underexpressed depending on the stress [36]. As for the candidate gene coding for the Hydroxyproline-rich glycoprotein-like identified in the QTLs detected in the dry and cold environments: RF, INC, and EP (Supplementary: Table S1), this gene was associated with drought tolerance in barley [37] and with the resistance to bacterial pathogen in rice [38].
Further, other candidate genes identified in the dry environment (RF): GDSL esterase/lipase EXL3; the cold environment (EP): UDP-glycosyltransferase 79-like isoform, and the glutathione S-transferase identified in both environments (Supplementary: Table S1). These genes play role in plant development and growth through the action on ABA homeostasis [39]; germination and growth [40]; detoxification and plant pathogen resistance [41].
4.1.2. Cold environment
In the cold environment (EP), among the candidate genes composing the QTL detected: 5′-3′ exoribonuclease 3 isoform X1, NADH dehydrogenase; as for the QTL of dry environment (RF), it contains the genes: starch synthase3, chloroplastic/amyloplastic and a photosystem I P700 chlorophyll an apoprotein A1 candidate gene. While the gene coding for the Photosystem II 44 kDa protein were identified in the QTLs for the cold (EP) and dry (RF) environment, as for the ankyrin repeat family protein-like it was found in the INC environment (Supplementary Table S1). All these candidate genes expression was altered under water stress [30]. Some candidate genes identified in QTLs for INC and RF environments like zinc finger family genes play an important role in physiological traits of yield potential under heat stress [27]. Moreover, in the QTL for the RF environment, the gene coding for BTB/POZ and MATH domain-containing protein 2 was highly repeated in this QTL, this gene is involved in heat tolerance and in plant growth and development [42].
4.1.3. Hot environment
In the hot environment RQ, six candidate genes were identified, one coding for LRR receptor-like serine/threonine-protein kinase RCH1 involved in resistance to pathogens [43, 44]. The remaining candidate genes were belonging to the family of retrotransposon/transposase. This gene family was also present in the other QTLs especially in RF (dry environment) with a high frequency. The abundance of this gene family presence can be explained by their involvement in stress conditions, [30] reported that the activity of these genes was altered under water stress and by the fact that the retrotransposon/transposon may represents 80 to 90% of wheat genome [45].
4.2. Detection of candidate genes in other chromosomes
Additionally, the presence of the wsnp_993293 SNP marker in the chromosome 1A and its linkage with the candidate gene implicated in powdery mildew resistance, confirms earlier mention on one parent of the population (Lahn) that carries moderate resistance to powdery mildew. Additionally, the wsnp_2265986, located in the 1B chromosome and linked to 3-ketoacyl-CoA synthase 6-like, this candidate gene plays also a role in abiotic (drought, heat and cold) and in biotic stress resistance. This reflects the genes carried by the second parent Cham1 for good drought and heat tolerance, osmotic adjustment, and proline content.
The candidate genes covered the entire genome and were linked to different biotic (powdery mildew, rust, etc.) and abiotic (drought, cold, heat, etc.) stress resistance / tolerance (Fig 4).
In the current work, a large part of candidate genes harbored by the 2A, in addition to 4B chromosome, were related to stress tolerance. They were also involved in plant growth and development. The 4BS results corroborate earlier studies [25, 46-50] on the importance of the 4BS and its contribution to grain yield in stressed environments.
5. Conclusion
The candidate genes found were linked to yield performance under drought and temperature extreme conditions. Also, this study showed the importance of 2A chromosomes for contrasting environments (drought, cold, and heat).
In this study specific and general candidate genes were detected. For the specific environments, the candidate genes in cold environment (EP) involved the genes of 5′-3’exoribonuclease 3 isoform X1 and NADH dehydrogenase. As for the dry environment (RF) the genes included were starch synthase3, chloroplastic/amyloplastic, and a photosystem I P700 chlorophyll. Further, in the dry environment (RF), the gene coding for BTB/POZ and MATH domain-containing protein2 was highly repeated in this environment. This gene is also effectuates heat tolerance, plant development and growth. Similarly, the ankyrin repeat family protein-like was found in the dry environment (INC), it contains the genes synthase3, chloroplastic/amyloplastic and photosystem I P700 chlorophyll. For the general candidate genes involved in diverse stressed environments, the following genes were identified: The Photosystem II 44 kDa protein were identified in the cold (EP) and the dry (RF) environments. Also, some of the candidate genes are operating in all 3 stresses, such as the zinc finger family genes. This gene also plays a significant role in physiology of yield potential under heat stress frequently.
5.1.1. Conflict of Interest
The authors declare no conflict of interest.
5.1.2. Acknowledgement
We would like to thank the International Center for Agricultural Research in Dry Area (ICARDA) for the financial support of this study and the support staff of the CIMMYT/ICARDA durum-breeding program.
- I. Elouafi, MM. Nachit, “A genetic linkage map of the Durum x Triticum dicoccoides backcross population based on SSRs and AFLP markers, and QTL analysis for milling traits” Theor Appl Genet, 108(3), 401-413, 2004. https://doi.org/10.1007/s00122-003-1440-8
- M.M. Nachit, “Durum wheat breeding for Mediterranean drylands of North Africa and West Asia. In: Rajram S, Saari EE, Hetel GP (eds) Durum Wheats” Challenges and Opportunities. CIMMYT, Ciudad Obregon, Mexico, 14, 14–27, 1992.
- El Yadini, I. Kahama, M. Labhilili, A. El Yadini, M. Azeqour, “La technique TILLING comme nouvelle biotechnologie pour la protection et l’amélioration du blé dur (TILLING : A new biotechnological method for protection and improvement of durum wheat)” J Mater Env Sci, 4, 931-934, 2013.
- M. Maccaferri, M. Sanguineti, S. Corneti, J. Ortega, M. Salem, J. Bort, E. DeAmbrogio, L. del Moral, A. Demontis, A. El-Ahmed, “Quantitative trait loci for grain yield and adaptation of durum wheat (Triticum durum Desf.) across a wide range of water availability” Genetics, 178(1), 489-511, 2008.
https://doi.org/10.1534/genetics.107.077297 - B. Heidari, BE. Sayed-Tabatabaei, G. Saeidi, M. Kearsey, K. Suenaga, P. Gulick, “Mapping QTL for grain yield, yield components, and spike features in a doubled haploid population of bread wheat” Genome, 54(6), 517-527, 2011. https://doi.org/10.1139/g11-017
- A. Blanco, M. Bellomo, A. Cenci, C. De Giovanni, R. D’ovidio, E. Iacono, B. Laddomada, M. Pagnotta, E. Porceddu, A. Sciancalepore, “A genetic linkage map of durum wheat” Theor App Genet 97(5-6), 721-728, 1998. https://doi.org/10.1007/s001220050948
- S.A. Dura, MA. Duwayri, MM. Nachit, “Detection of molecular markers associated with yield and yield components in durum wheat (Triticum turgidum L. var. durum Desf.) under drought conditions” Afr J Agr, 8(19), 2118-2128, 2013. https://doi.org/10.5897/AJAR11.2031
- MM. Nachit, I. Elouafi, A. Pagnotta. A. El saleh, E. acono, M. Labhilili, A. Asbati, M. Azrak, H. azzam, D. Benscher, M. Khairallah, JM. Ribaut, OA. Tanzarella, E. Porceddu, ME. Sorrells, “Molecular linkage map for an intraspecific recombinant inbred population of durum wheat (Triticum turgidum L. var. durum)”. Theor App Genet, 102, 177-186, 2001. https://doi.org/10.1007/s001220051633
- A. Alsaleh, FS. Baloch, M. Derya, M. Azrak, B. Kilian, H Özkan, MM. Nachit, “Genetic Linkage Map of Anatolian Durum Wheat Derived from a Cross of Kunduru-1149× Cham1”. Plant Mol Biol Rep, 33, 209-220, 2014. https://doi.org/10.1007/s11105-014-0749-6
- Mantovani, M. Maccaferri, MC. Sanguineti, R. Tuberosa, I. Catizone, P. Wenzl, B. Thomson, J. Carling, E. Huttner, E. DeAmbrogio, “An integrated DArT-SSR linkage map of durum wheat” Mol Breed, 22(4), 629-648, 2008.https://doi.org/10.1007/s11032-008-9205-3
- M. Nagel, S. Navakode, V. Scheibal, M. Baum, MM. Nachit, M. Röder, A. Börner, “The genetic basis of durum wheat germination and seedling growth under osmotic stress” Biol Plantarum, 58, 681–688, 2014.
https://doi.org/10.1007/s10535-014-0436-3 - KJ. Haile, MM. Nachit, H. Karl, B. Ayele, SM. Röder, “QTL mapping of resistance to race Ug99 of Puccinia graminis f. sp. tritici in durum wheat (Triticum durum Desf.)” Mol Breed, 30, 1479–1493, 2012. https://doi.org/10.1007/s11032-012-9734-7
- S. Dura, M. Duwayri, MM. Nachit, F. Al Sheyab, “Detection of molecular markers associated with yield and yield components in durum wheat (Triticum turgidum L. var. durum) under saline conditions” Crop Past Sci, 64(10), 957-964, 2013. https://doi.org/10.5897/AJAR11.2031
- RM. Poecke, M. Maccaferri, J. Tang, HT. Truong, A. Janssen, NJ. Orsouw, S. Salvi, MC. Sanguineti, R. Tuberosa, EA Vossen, “Sequence based SNP genotyping in durum wheat” Plant Biotech J, 11(7), 809-817, 2013. https://doi.org/10.1111/pbi.12072
- D. Trebbi, M. Maccaferri, P. de Heer, A. Sørensen, S. Giuliani, S. Salvi, M. Sanguineti, A. Massi, E. van der Vossen, R. Tuberosa, “High-throughput SNP discovery and genotyping in durum wheat (Triticum durum Desf.)” Theor App Genet, 123(4), 555-569, 2011.
https://doi.org/10.1007/s00122-011-1607-7 - W. Zhang, S. Chao, F. Manthey, O. Chicaiza, JC. Brevis, V. Echenique, J. Dubcovsky, “QTL analysis of pasta quality using a composite microsatellite and SNP map of durum wheat” Theor App Genet, 117(8) 1361-1377, 2008.https://doi.org/10.1007/s00122-008-0869-1
- A. Blanco, P. Colasuonno, A. Gadaleta, G. Mangini, A. Schiavulli, R. Simeone, AM. Digesù, P. De Vita, AM. Mastrangelo, L. Cattivelli, “Quantitative trait loci for yellow pigment concentration and individual carotenoid compounds in durum wheat” J Cereal Sci, 54(2), 255-264, 2011.
https://doi.org/10.1016/j.jcs.2011.07.002 - A. Blanco, G. Mangini, A. Giancaspro, S. Giove, P. Colasuonno, R. Simeone, A. Signorile, P. Vita, AM. Mastrangelo, L Cattivelli, “Relationships between grain protein content and grain yield components through quantitative trait locus analyses in a recombinant inbred line population derived from two elite durum wheat cultivars” Mol Breed, 30(1), 79-92, 2011. https://doi.org/10.1007/s11032-011-9600-z
- A. Gadaleta, A. Giancaspro, S. Giove, S. Zacheo, G. Mangini, R. Simeone, A. Signorile, A. Blanco, “Genetic and physical mapping of new EST-derived SSRs on the A and B genome chromosomes of wheat” Theor App Genet, 118(5), 1015-1025, 2009. https://doi.org/10.1007/s00122-008-0958-1
- D. Marone, G. Laidò, A. Gadaleta, P. Colasuonno, D. Ficco, A. Giancaspro, S. Giove, G. Panio, M. Russo, P. De Vita, L. Cattivelli, R. Papa, A. Blanco, “A high-density consensus map of A and B wheat genomes” Theor App Genet, 125(8), 1619-1638, 2012. https://doi.org/10.1007/s00122-012-1939-y
- MA. Russo, DBM. Ficco, D. Marone, P. De Vita, V. Vallega, C. Rubies-Autonell, C. Ratti, P. Ferragonio, V. Giovanniello, N. Pecchioni, “A major QTL for resistance to soil-borne cereal mosaic virus derived from an old Italian durum wheat cultivar” J Plant Interact, 7(4), 290-300, 2012.
https://doi.org/10.1080/17429145.2011.640437 - J. Ahmad et al., “Study of Physio-chemical Properties of POSS/Mineral Oil based Nanofluids,” in 2018 International Conference on Power Generation Systems and Renewable Energy Technologies (PGSRET), 1–5, 2018. DOI: 10.1109/PGSRET.2018.8685972
- Z. Peleg, Y. Saranga, T. Suprunova, Y. Ronin, MS. Röder, A. Kilian, AB. Korol, T. Fahima, “High-density genetic map of durum wheat x wild emmer wheat based on SSR and DArT markers” Theor App Genet, 117, 103–115, 2008. https://doi.org/10.1007/s00122-008-0756-9
- H. Tura, J. Edwards, V, Gahlaut, M. Garcia, Sznajder, U. Baumann, F. Shahinnia, M. Reynolds, P. Langridge, HS. Balyan, “QTL analysis and fine mapping of a QTL for yield-related traits in wheat grown in dry and hot environments” Theor App Genet, 1-19, 2019.
https://doi.org/10.1007/s00122-019-03454-6 - D. Xu, W. Wen, L. Fu, F. Li, J. Li, L. Xie, X. Xia, Z. Ni, Z. He, S. Cao, “Genetic dissection of a major QTL for kernel weight spanning the Rht-B1 locus in bread wheat” Theor App Genet, 132(11), 3191-3200, 2019. https://doi.org/10.1007/s00122-019-03418-w
- R. Mérida-García, G. Liu, S. He, V. Gonzalez-Dugo, G. Dorado, S. Gálvez, I. Solís, P. Zarco-Tejada, J. Reif, P. Hernandez P, “Genetic dissection of agronomic and quality traits based on association mapping and genomic selection approaches in durum wheat grown in Southern Spain” PloS one 14(2), e0211718, 1-24, 2019. https://doi.org/10.1371/journal.pone.0211718
- S. Pradhan, MA. Babar, G. Bai, J. Khan, D. Shahi, M. Avci, J. Guo, J. McBreen, S. Asseng, S. Gezan, et al, “Genetic dissection of heat-responsive physiological traits to improve adaptation and increase yield potential in soft winter wheat” BMC Genomics 21, 315, 1-15, 2020.
https://doi.org/10.1186/s12864-020-6717-7 - M.S. Rahman, K.J. Linsell, J.D. Taylor, M.J. Hayden, N.C. Collins, K.H. Oldach, “Fine mapping of root lesion nematode (Pratylenchus thornei) resistance loci on chromosomes 6D and 2B of wheat” Theor App Genet 133, 635-652. 2020.
https://doi.org/10.1007/s00122-019-03495-x - J. Yu, Y. Miao, S. Yang, Z. Shi, N. Miao, M. Ding, H. Zhang, Y. Jiang, J. Rong, “Identification and mapping of a photoperiod response gene (QPpd.zafu-4A) on wild emmer wheat (Triticum turgidum L.) chromosome 4AL” Euphytica 215, 146, 1-12, 2019.
https://doi.org/10.1007/s10681-019-2469-3 - D. Habash, M. Baudo, M. Hindle, S. Powers, M. Defoin-Platel, R. Mitchell, M. Saqi, C. Rawlings, K. Latiri, J. Araus, et al, “Systems responses to progressive water stress in durum wheat” PloS one, 9, e108431, 1-21, 2014. https://doi.org/10.1371/journal.pone.0108431
- I. Farouk, F. Gaboun, Z. Kehel, A. Alsaleh, B. Belkadi, I. Elouafi, J. Motowaj, A. Filali maltouf, MM. Nachit, “Dissection of QTL linked to grain yield and identification of candidate genes involved in grain yield formation using comparative SNP sequences analysis,” in 2020 IEEE the first International Conference on Innovative Research in Applied Science, Engineering and Technology (IRASET), Meknes, Morocco, pp. 1-6, 2020.
https://doi.org/10.1109/IRASET48871.2020.9091989 - S. Li, J. Jia, X. Wei, X. Zhang, L. Li, H. Chen, Y. Fan, H. Sun, X. Zhao, T. Lei, et al, “A intervarietal genetic map and QTL analysis for yield traits in wheat” Mol Breeding 20, 167-178, 2007. https://doi.org/10.1186/s12870-018-1308-3
- S. Sukumaran, M.P. Reynolds, C. Sansaloni, “Genome-Wide Association Analyses Identify QTL Hotspots for Yield and Component Traits in Durum Wheat Grown under Yield Potential, Drought, and Heat Stress Environments”. Front Plant Sci 9, 81, 1-16, 2018.
https://doi.org/10.3389/fpls.2018.00081 - P. Colasuonno, MA. Maria, A. Blanco, Gadaleta, “Description of durum wheat linkage map and comparative sequence analysis of wheat mapped DArT markers with rice and Brachypodium genomes” BMC Genet, 14, number (114), 1-9, 2013. https://doi.org/10.1186/1471-2156-14-114
- C. Xin, R. Hou, F. Wu, Y. Zhao, H. Xiao, W. Si, ME. Ali, L. Cai, J. Guo, “Analysis of cytosine methylation status in potato by methylation-sensitive amplified polymorphisms under low-temperature stress” J Plant Biol, 58, 383-390, 2015. https://doi.org/10.1007/s12374-015-0316-1
- M. Jain, A. Nijhawan, R.Arora, P. Agarwal, S. Ray, P. Sharma, S. Kapoor, AK. Tyagi, JP. Khurana, “F-Box Proteins in Rice. Genome-Wide Analysis, Classification, Temporal and Spatial Gene Expression during Panicle and Seed Development, and Regulation by Light and Abiotic Stress” Plant Physiol, 143, 1467-1483, 2007. https://doi.org/10.1104/pp.106.091900
- SF. Abou-Elwafa, “Identification of genes associated with drought tolerance in barley” Biol Plantarum 62, 299-306. 2018.
https://doi.org/10.1007/s10535-017-0765-0 - IS. Kumar, N. Zaharin, K. Nadarajah, “In silico identification of resistance and defense related genes for bacterial leaf blight (BLB) in rice” J Pure Appl Microbiol, 12, 1867-1877, 2018. https://dx.doi.org/10.22207/JPAM.12.4.22
- Z. Liu, J-P. Yan, D-K. Li, Q. Luo, Q. Yan, Z-B. Liu, L-M. Ye, J-M. Wang, X-F. Li, Y. Yang, “UDP-Glucosyltransferase71C5, a Major Glucosyltransferase, Mediates Abscisic Acid Homeostasis in Arabidopsis” Plant Physiol, 167, 1659-1670, 2015. https://doi.org/10.1104/pp.15.00053
- C-P. Lai, L-M. Huang, L-FQ. Chen, M-T. Chan, J-F. Shaw, “Genome-wide analysis of GDSL-type esterases/lipases in Arabidopsis” Plant Mol Biol, 95, 181-197, 2017. https://doi.org/10.1007/s11103-017-0648-y
- G. Gullner, T. Komives, L. Király L, P. Schröder, “Glutathione S-Transferase Enzymes in Plant-Pathogen Interactions” Front Plant Sci 9, 1836, 1-19, 2018. https://doi.org/10.3389/fpls.2018.01836
- K. Morimoto, N. Ohama, S. Kidokoro, J. Mizoi, F. Takahashi, D. Todaka, J. Mogami, H. Sato, F. Qin, J-S. Kim, et al, “BPM-CUL3 E3 ligase modulates thermotolerance by facilitating negative regulatory domain-mediated degradation of DREB2A in Arabidopsis” Proceedings of the National Academy of Sciences 114(40), E8528-E8536, 2017. https://doi.org/10.1073/pnas.1704189114
- S. Padmarasu, DJ. Sargent, A. Patocchi, M. Troggio, P. Baldi, G. Linsmith, L. Poles, M. Jänsch, M. Kellerhals, S. Tartarini, R. Velasco, “Identification of a leucine-rich repeat receptor-like serine/threonine-protein kinase as a candidate gene for Rvi12 (Vb)-based apple scab resistance” Mol Breeding, 38 ,73, 1-14, 2018. https://doi.org/10.1007/s11032-018-0825-y
- D. Pradhan, D. Mathew, SK. Mathew, PA. Nazeem, “Identifying the markers and tagging a leucine-rich repeat receptor-like kinase gene for resistance to anthracnose disease in vegetable cowpea [Vigna unguiculata (L.) Walp.]” J Hortic Sci Biotech, 93, 225-231, 2018. https://doi.org/10.1080/14620316.2017.1362962
- V. Jamilloux, J. Daron, F. Choulet, H. Quesneville, “De novo annotation of transposable elements: tackling the fat genome issue” Proceedings of the IEEE, 105, 474-481, 2016. https://10.1109/JPROC.2016.2590833
- AA. Diab, R. Kantety, N. Ozturk, D. Benscher, MM. Nachit, M. Sorrells, “Drought-inducible genes and differentially expressed sequence tags associated with components of drought tolerance in durum wheat” Sci Res Essay, 3, 009-026, 2008.
- D. Habash, Z. Kehel, M. Nachit, “Genomic approaches for designing durum wheat ready for climate change with a focus on drought” J Exp Bot, 60(10), 2805-2815, 2009. https://doi.org/10.1093/jxb/erp211
- MM. Nachit, “Association of grain yield in dryland and carbon isotope discrimination with molecular markers in durum (Triticum turgidum L. var. durum)” In Proceedings of 9th International Wheat Genetics Symposium, Saskatoon, ed. A.E. Slinkard, Canada, 218–223, 1998.
- MM. Nachit, I.Elouafi, “Durum wheat adaptation in the Mediterranean dryland: Breeding, stress physiology, and molecular markers” In Challenges and strategies for dry-land agriculture, eds. S.C. Rao and J. Ryan, CSSA. Madison, WI: Crop Science of America and American Society of Agronomy, 32, 203–218, 2004. https://doi.org/10.2135/cssaspecpub32.c13
- J. Li, S. Wen, C. Fan, M. Zhang, S. Tian, W. Kang, W. Zhao, C. Bi, Q. Wang, S. Lu, et al, “Characterization of a major quantitative trait locus on the short arm of chromosome 4B for spike number per unit area in common wheat (Triticum aestivum L.)” Theor Appl Genet, 133, 2259–2269, 2020.
https://doi.org/10.1007/s00122-020-03595-z

