Towards Classification of Shrimp Diseases Using Transferred Convolutional Neural Networks
Towards Classification of Shrimp Diseases Using Transferred Convolutional Neural Networks
Volume 5, Issue 4, Page No 724-732, 2020
Author’s Name: Nghia Duong-Trung1, Luyl-Da Quach1, Chi-Ngon Nguyen2,a)
View Affiliations
1Software Engineering Department, FPT University, Can Tho city, 94000, Vietnam
2College of Engineering Technology, Can Tho University, Can Tho city, 94000, Vietnam
a)Author to whom correspondence should be addressed. E-mail: ncngon@ctu.edu.vn
Adv. Sci. Technol. Eng. Syst. J. 5(4), 724-732 (2020); ![]() DOI: 10.25046/aj050486
DOI: 10.25046/aj050486
Keywords: Shrimp diseases, Transfer learning, Convolutional neural networks
Export Citations
Vietnam is one of the top 5 largest shrimp exporters globally, and Mekong delta of Vietnam contributes more than 80% of total national production. Along with intensive farming and growing shrimp farming area, diseases are a severe threat to productivity and sustainable development. Timely response to emerging shrimp diseases is critical. Early detection and treatment practices could help mitigate disease outbreaks, leading to on-site diagnostics, instant services recommendation, and front-line treatments. The authors establish a con- tribution hub for data collection in the ethnographic fieldwork of Mekong delta. Several deep convolutional neural networks are trained by applying the transfer learning technique. We have investigated six common reported shrimp diseases. The classification accuracy is achieved of 90.02%, which is very useful in extremely non-standard images. Throughout the work, we raise the attention of shrimp experts, computer scientists, treatment agencies, and policymakers to develop preventive strategies against shrimp diseases.
Received: 29 May 2020, Accepted: 12 August 2020, Published Online: 28 August 2020
1. Introduction
The shrimp farming industry in Vietnam has been expanding continuously for decades, especially in the Mekong Delta. Regarding the shrimp industry alone, the region contributes more than 80% of total production, mainly from Bac Lieu (BL), Ca Mau (CM), Ben Tre (BT), and Soc Trang (ST) [1], see Figure 1. In 2017, these provinces contributed more than 50% of Vietnam’s total shrimp production. In 2018, the region’s total shrimp farming area was about 720,000 hectares, with a total of 745,000 tons. Leading the production is the giant tiger shrimp (Penaeus monodon), whose output is around 300 ktons per year. In the first seven months of 2019, Vietnam’s shrimp exports reached US$ 1.8 billion (https://tongcucthuysan.gov.vn/en-us/Fisheries-Trading/doctin/013419/2019-08-27/vietnam-shrimp-exports-started-toreverse). The leading shrimp importers of Vietnam include the EU, the USA, Japan, China, Korea, Canada, Australia, ASEAN countries, and Switzerland, accounting for 96% total export value (http://seafood.vasep.com.vn/702/onecontent/certifications.html).
On January 18, 2018, Decision 79/QD-TTg of the Vietnamese Prime Minister issued a national plan for shrimp industry development to 2025. The goal is to export shrimp worth 10 billion USD from 2020 to 2025 (https://customsnews.vn/small-productionhindered-vietnamese-shrimp-industry-10734.html) (i.e., an average growth of 12.7% per year) of which the value of brackish water shrimp exports will be USD 8.4 billion; the total area of brackish shrimp farming will reach 750,000 hectares; the centralized area for rearing giant freshwater prawn will be 50,000 hectares; lobster farming will reach 1,300,000m3 cages; total shrimp production will reach 1,153,000 tons (average growth of 6.73% per year); brackish shrimp will be archived by 1,100,000 tons; giant freshwater shrimp reaches 50,000 tons, and lobster reaches 3,000 tons (http://agro.gov.vn/vn/tID25840 Nam-2020-xuat-khau-tomdat-55-ty-USD.html). Although they occupy less than 10% of the land devoted to shrimp farming, they contribute about 80% of total production [?]. The growth of the shrimp industry is necessary to provide shrimp-based products for increasing global population demands. The need for such manageable growth is intensively recognized as an essential goal. However, strengthening the intensification of production can also make shrimp more susceptible to diseases. For example, in 2012, one-sixth of the total area was severely damaged by infectious diseases, acute hepatopancreatic necrosis disease (AHPND), or early mortality syndrome (EMS) [2]. Warnings on potentially sustainable intensification indicate the increasing understanding of the government and shrimp farmers of the environmental, economic, and social impacts of increasing aquaculture production and its trade-off. The Vietnamese government has cooperated with international partners to find solutions to improve the sustainable and effective shrimp farming practices of the country and the Mekong Delta, focusing on small-scale shrimp farming (https://psmag.com/environment/the-environmentalimpacts-of-shrimp-farming-in-vietnam), see Figure 2. Along with intensive farming and growing shrimp farming area, diseases are a severe threat to productivity and sustainable development of shrimp farming.
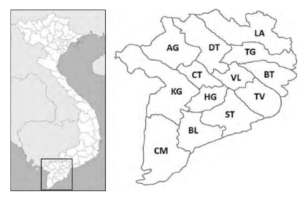
Figure 1: Mekong Delta of Vietnam and its provinces.
Although there are many policies and plans from the government and localities for the development of the shrimp industry in the Mekong Delta, shrimp production in many provinces continues to suffer significant economic losses due to the impact of shrimp diseases. Besides, the intensive farming of many shrimp crops per year is also affected because shrimp farmers have to spend time preparing new shrimp ponds after the shrimp diseases. According to an FAO survey conducted from 2013 to 2016, all respondents said shrimp diseases affected their farms (http://www.fao.org/3/ca6702en/ca6702en.pdf). Commonly reported diseases are AHPND, EMS, white spot syndrome virus (WSSV), white feces syndrome (WFS), and yellow-headed virus (YHV). Regarding WSSV, shrimp farmers are aware of an infection occurrence over an extended period, e.g., from 25 to 60 days after stocking. The delay in disease detection and treatment intervention can lead to the severe loss of the entire shrimp crop. Shrimp farmers reported that EMS, WSSV, and YHV diseases could not be controlled while WFS is manageable. The most regular exercise shrimp farmers apply to reduce disease risks is to buy quality food from reputable hatcheries and prepare and disinfect ponds. However, these reactions depend significantly on their own experiences without much scientific knowledge and standardization of treatment. These exercises make the shrimp industry in Mekong Delta vulnerable to the rapid and un-manageable propagation of emerging diseases. According to statistics on Asian Fisheries Science, 8.72% of Penaeus monodon ponds and 32.48% of Penaeus vannamei ponds in 2014 were reported to have been affected by shrimp disease [3]. In 2015, the monodon-affected ponds of AHPND were reported to be 5,875 hectares, while more than 5,509 hectares used for Penaeus vannamei culture were infected. WSSV remains the most important viral pathogen of farmed shrimp, with the ability to infect quickly and often results in from 80% to 100% crop failure. Infections of Penaeus monodon and Penaeus vannamei ponds in the Mekong Delta in 2015 were estimated US$ 11.02 million loss. When preparing to submit this article, the authors read the information in Quang Ngai province (Vietnam) and knew that the People’s Committee of Binh Son district had just decided to handle the compulsory destruction of 21 diseased shrimp ponds (http://vietlinh.vn/tin-tuc/2020/nuoitrong-thuy-san-2020-s.asp?ID=470). Accordingly, local authorities were forced to destroy all white leg shrimp in 21 shrimp ponds with an area of 49700m2 of 11 shrimp farmers. Dead shrimps were diagnosed with WSSV and AHPND. Shrimp farmers did not know the identity of shrimp diseases and asked for information on shrimp forums. It causes devastating delays in detection and timely intervention.
Consequently, timely response to emerging shrimp diseases is critical. Early detection and treatment practices could help mitigate disease devastation, leading to on-site diagnostics, instant services recommendation, and front-line treatments.
In contrast with the abundance of research on shrimp diseases [4]- [7], computer science applications for them are uncommonly explored, especially identification and classification. Thanks to the ubiquity of mobile devices and social media networks, shrimp farmers have many online channels and forums to post images of shrimp diseases and ask for any detection, help on knowledge and treatments, see two examples in Figure 3. The volume of information related to shrimp diseases has been accumulated overtimes, and those being generated by shrimp farmers should be notified to experts, treatment agencies, and policymakers. There is a need to have a place to collect images for integrating legacy information with contemporary knowledge related to shrimp diseases. Accurate and early detection is preliminary to any intervention and recommendation of treatments. It is obvious to say that early detection and intervention are better than treatment when diseases get worse. Hence, leveraging technology in the easily automatic identification of these diseases has become essential.

Figure 2: An example of small-scale shrimp farming in Mekong Delta. Shrimp farmers dig a pond and establish several holding pens next to their house. Image courtesy to Roberto Schmidt/AFP/Getty Images.
To the best of our knowledge, the authors have made several contributions as follows.
- First, we establish a contribution hub where farmers can send images of emerging shrimp diseases. This kind of contribution hub or center has not existed before. Several images have been sent to shrimp experts at local universities for labeling. • Then, a machine learning system is deployed to identification diseases. Although the number of images is small, the detection accuracy should not be sacrificed and less demand for computing power. More specifically, the authors apply convolutional neural networks applying the transfer learning technique. Addressing those requirements lead to our second contribution.
- Third, we have investigated six common reported shrimp diseases, which make the experiments the most intensive in the literature.
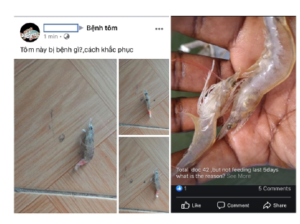
Figure 3: Shrimp farmers took pictures of infected samples, posted them to a Facebook forum and asked for help on the emerging diseases. The text above the pictures on the left means ”What is the disease of this shrimp? How to cure?”
2. Related Work
The purpose of this work is neither to discuss existing research on shrimp diseases in aquaculture through the lens of biologists [8, 9] nor the effects of diseases over shrimp industry from an ecological perspective [10].By the time we wrote a literature review, there is very little research on the classification or detection of shrimp diseases based on images. YHV recognition application was designed in [11]. They proposed an image processing algorithm based on the Niblack algorithm to detect and eliminate shrimp with only YHV from the gather lines. Another work devoted to WSSV detection in shrimp images using the K-means clustering technique (https://www.semanticscholar.org/paper/WhiteSpot-Syndrome-Virus-Detection-in-Shrimp-using-NagalakshmiJyothi/5d77b95d947adf375139a01662b6b308e2024811). They equipped special spectrographic cameras, which are either not affordable to shrimp farmers or applicable in real scenarios. Another exciting research on the identification of softshell and sound shrimps, e.g., caused by physical illness or pathological disease, has been done in [12]. Several exciting shrimp image analysis applications but not disease detection can be referred to as shrimp freshness evaluation [13] and marine organism classification [14].
Many previous researchers have focused their attention on deploying a more accurate deep convolutional neural network regarding shrimp images. Zihao et al. developed ShrimpNet-3 [15], for recognizing shrimp, which is an architecture modification from LeNet-5 [16]. Similarly, the idea of modifying the architecture of a deep neural network like AlexNet, in this case, is investigated in [17]. That paper addressed the task of classification between a softshell and sound shrimp from an imaging perspective. The mentioned works examined several architectures, but the main point is that the networks need to be trained from scratch. These papers have not investigated the transfer learning approach which our work intends to.
3. System Components
In this Section, the authors briefly describe the proposed architecture to satisfy the paper’s contributions. We illustrate our system components in Figure 4. We launch a data collection hub[1] where shrimp farmers across Mekong Delta region and other countries can send their images. It happens is step 1 in the system components. In step 2, shrimp images are sent to experts in local universities for ground-truth labeling. We can see in Figure 3, shrimp farmers send pictures with a diverse background, including fingers, catching equipment, soil, and floor. In step 3, we replace the background by white color. Steps 4, 5, and 6 are the necessary machine learning process. However, we do not train the models from scratch. Instead, we apply the transfer learning technique, which is further described in Section (4). At step 7, we contact the farmers who had sent shrimp images to ask for further details of the emerging diseases.
4. Technical Background
4.1 Transfer Learning
The traditional machine learning process emphasizes the entire process of independent learning from data collection, model selection and training, performance evaluation, and metadata tuning. Modern object identification and classification models with millions of parameters can take weeks to complete training. Therefore, the first need is that the database used for model training must be enormous. The second requirement is computer infrastructure and financial support, especially in high power-consumption algorithms such as neural networks.
Definition 1 Given a training data set Xtrain ∈ Rn×m and equivalent label Ytrain ∈ Rn where n, and m are the number of samples and the number of attributes, respectively. Traditional machine learning methods seek to build a f prediction model by training on pairs (Xtrain,Ytrain) = {(xi,yi),…,(xn,yn)}.
A principal assumption in applying machine learning algorithms is that the training and test data must share a similar feature space, more specifically Xtrain and Xtest are described using the same attributes. However, this assumption can be difficult to satisfy in many real-world applications. It reduces the generalization possibilities of machine learning models. The difference between transfer learning traditional machine learning is shown in Figure 5.
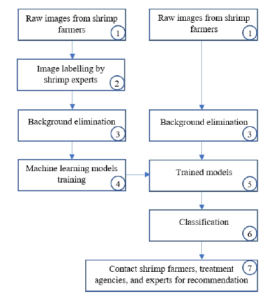
Figure 4: Our proposed system components.
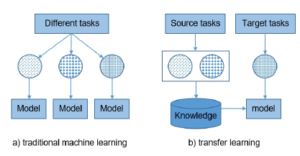
Figure 5: Difference between traditional machine learning and transfer learning.
The idea of transferring knowledge is a new approach to machine learning practices. It develops a mechanism of knowledge transferability in one or more source tasks and uses it to improve the prediction capacity in a new task [18]- [21]. It is like the propagation of knowledge from a well-developed domain with a lot of learning data to a less-developed domain that is limited due to insufficient data [22, 23]. The method allows machine learning models to be applied to new data drawn from distributions that are entirely different from the original data sources. Machine learning algorithms avoid suffering from the cold start problems by leveraging fully trained models on predefined large datasets [24, 25], Transfer learning aims to improve generalization ability in the target task by leveraging knowledge from the source task(s). We can define transfer learning as follows.
Definition 2 Given a source domain Dsource where we define a learning task T source. We denote Dtarget and T target are a target domain and a target learning task, respectively. Transfer learning technique aims to help improve the learning of the f prediction model for T target utilizing knowledge in Dsource and T source, where Dsource , Dtarget or T source , T target.
By re-weighting the observations in Dsource, the effects of the various samples are reduced. Conversely, similar instances will contribute more to T target and may lead to a more accurate prediction.
4.2 Convolutional Neural Networks (CNNs)
Convolutional neural networks (CNNs), initially attending several large-scale image classification challenges [24]-[26], is a class of deep, feed-forward artificial neural networks that are applied to the vast majority of machine learning problems due to the outstanding performance. The architecture of CNNs is usually a composition of layers that can be grouped by their functionalities. The high performance of CNNs is achieved by (i) their ability to learn rich level image representations and (ii) leveraging a tremendous amount of data. It can take millions of estimated parameters to characterize the network. However, in the situation of limited data sources, training CNNs from scratch is not very useful. There is a phenomenon in the learning of level image that each layer represents from color blobs, lines, and basic shapes to a high-level mixture of them. Several first-layer patterns appear to be generic to any dataset, Hense, its knowledge can be transferred and re-used. CNNs can be trained on large-scale datasets, and then re-weighted on a particular target dataset. The idea of training deep neural networks applying transfer learning has been explored by previous research [27]-[30].
4.2.1 Inception-based models
The Inception deep convolutional architecture was introduced as
GoogLeNet or Inception-v1 in [31]. Later architecture improvement was the introduction of batch normalization or Inception-v2 [32] and additional factorization or Inception-v3 [33].
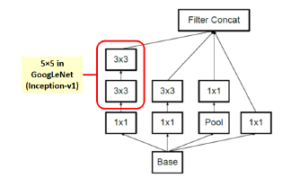
Figure 6: Inception module A.
In image representation, the prominent patterns can exist in different size variations. Therefore, the problem is to choose the appropriate filter size that slides through the image. The idea of inception effectively addresses the mentioned problem by introducing several Inception modules where multiple-size filters operate on the same level. See Figures 6, 7, and 8 or Inception Module A, B, and C respectively. Consequently, the computational expense is significantly reduced because of the reduction of connections/parameters. The outline of our adapted implementation architecture is described in Table 10.
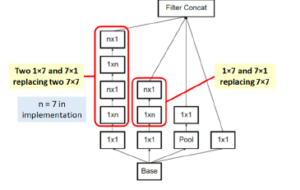
Figure 7: Inception module B.
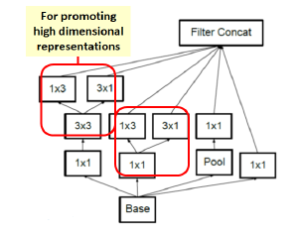
Figure 8: Inception module C.
4.2.2 Depthwise separable convolution-based model
MobileNets is a family of mobile-first computer vision models designed by Google [34]. Its primary design is to help develop robust applications on mobile devices where restricted resources exist. Similar to how other popular CNNs models, it can be developed for addressing classification, detection, and segmentation tasks. The main point in Mobilenets is that two separated depthwise and pointwise convolutions perform the filtering and combination processes. More precisely, the model splits the convolution into a 3 × 3 and a 1 × 1 depthwise and pointwise, respectively. The depth-wise convolution applies a single filter to each input channel; the pointwise convolution combines the outputs of the depth-wise ones. These two different convolutions are illustrated in Figure 9. The depthwise separable convolution architecture is described in Table 1. The comparison between the inception-v3 model and mobilenets is presented in Table 2.
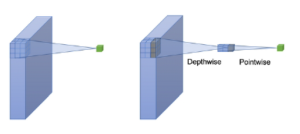
Figure 9: A depthwise and a pointwise convolutions.
Table 1: The MobileNets architecture [34]. At the softmax layer, c is the number of labels. dw means depthwise.
Layer (Stride) Filter size Input size
| conv2d (s2) | 3 × 3 × 3 × 32 | 224 × 224 × 3 |
| conv2d dw (s1) | 3 × 3 × 32 dw | 112 × 112 × 32 |
| conv2d (s1) | 1 × 1 × 32 × 64 | 112 × 112 × 32 |
| conv2d dw (s2) | 3 × 3 × 64 dw | 112 × 112 × 64 |
| conv2d (s1) | 1 × 1 × 64 × 128 | 56 × 56 × 64 |
| conv2d dw (s1) | 3 × 3 × 128 dw | 56 × 56 × 128 |
| conv2d (s1) | 1 × 1 × 128 × 128 | 56 × 56 × 128 |
| conv2d dw (s2) | 3 × 3 × 128 dw | 56 × 56 × 128 |
| conv2d (s1) | 1 × 1 × 128 × 256 | 28 × 28 × 128 |
| conv2d dw (s1) | 3 × 3 × 256 dw | 28 × 28 × 256 |
| conv2d (s1) | 1 × 1 × 256 × 256 | 28 × 28 × 256 |
| conv2d dw (s2) | 3 × 3 × 256 dw | 28 × 28 × 256 |
| conv2d (s1) | 1 × 1 × 256 × 512 | 14 × 14 × 256 |
| 5× conv2d dw (s1) conv2d (s1) |
3 × 3 × 512 dw 1 × 1 × 512 × 512 |
14 × 14 × 512 14 × 14 × 512 |
| conv2d dw (s2) | 3 × 3 × 512 dw | 14 × 14 × 512 |
| conv2d (s1) | 1 × 1 × 512 × 1024 | 7 × 7 × 512 |
| conv2d dw (s2) | 3 × 3 × 1024 dw | 7 × 7 × 1024 |
| conv2d (s1) | 1 × 1 × 1024 × 1024 | 7 × 7 × 1024 |
| avg pool (s1) | 7 × 7 | 7 × 7 × 1024 |
| fully connected (s1) | 1024 × 1000 | 1 × 1 × 1024 |
| softmax (s1) | predictions | 1 × 1 × c |
Table 2: The comparison between the inception-based and MobileNets models.
| Experimented models | # Mult-Adds (Million) | # Parameters (Million) |
| Inception | 5000 | 23.6 |
| MobileNets 1.0-244 | 569 | 4.24 |
5. Experiments
5.1 Datasets
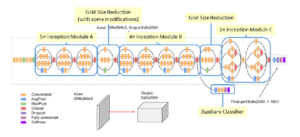
Figure 10: The outline of inception-based architecture. At the softmax layer, 1001 is replaced by the number of predicted labels in our dataset.
In this work, the authors have evaluated the machine learning models on different data sources. We collected images from a wide range of shrimp forums[2] [3] [4], where farmers posted the shrimp images and asked for help. We also launch a data collection hub as discussed in Section 3. The hub has been advertised to provinces administrations and shrimp farmers in Mekong Delta. The images were collected under various conditions, such as lighting, angle, distance, and background. Then, they were fed into learning models without manually resizing.
We sent raw images to experts at the College of Aquaculture & Fisheries[5] and experts in our project for labeling. The image background was manually removed using Adobe Photoshop by several technical assistants. Then the images were rechecked by the experts to be accepted as training images. Since the number of images we have collected weekly is not that much, the proposed manual pre-processing works acceptably.
Table 3: Shrimp diseases dataset used in the experiments.
# Label # Observations
| 1 | Black gill | 37 |
| 2 | Black spot | 52 |
| 3 | WSSV | 88 |
| 4 | IMNV | 45 |
| 5 | NHB | 47 |
| 6 | YHV | 35 |
| 7 | Healthy shrimp | 44 |
| Total images | 348 |
At the time of conducting this paper, the authors receive images of 6 most asked shrimp diseases such as YHV [35], black gill [36], black spot [37], WSSV [38], infectious myonecrosis virus (IMNV) [39], and necrotizing hepatopancreatitis bacterium (NHB) [40]. We also add healthy shrimps as the seventh class. Example of photos collected by the authors presented in Figure 11. We show the distribution of samples into classes in Table 3.
5.2 Experimental Results
The implementation process is executed using Anaconda Python Distribution for Python 3.6 installed in Windows 10, TensorFlow version 1.7, and NVIDIA GPU Computing Toolkit 10.2. Our experiments were conducted on a regular machine equipped with a Core i7 with 2.5GHz clock speed, GPU NVIDIA GeForce 940MX, and 16GB of RAM.
In our experiments, we grid-search several hyperparameters as follows. The learning rate (lr) is {0.1,0.01}. The number of epoch m(ep) is {1000,2000,3000}. We randomly shuffle the dataset into an 80% training set, a 10% validation set, and a 10% test set, which is a regular choice in machine learning practices. We refer to it as the 80/10/10 splitting schema. The models are tuned by the best score on the validation set. The tuning process is to find the best combination of hyperparameters of the learning rate and the number of epoch. After that, we run the train models on the test set for final classification accuracy. We run the experiments five times and take the average scores.
The input size of the MobileNets model is 224 × 224 × 3 for height, width, and channel, respectively. Regarding the Inception model, the input size is 299 × 299 × 3. The performance of models is reported in Table 4 and the identification accuracy is highlighted in Figure 12. The reported running time includes (i) shuffling images into 80/10/10 schema, (ii) training the model on the training/validation sets, and (iii) performing prediction capacity on the test set. Note that the best hyperparameters combination is 0.01 and 2000 for the learning rate and the number of epoch, respectively.

Figure 11: Shrimp diseases images. From left to right and the first row to bottom row, the labels are black gill, black spot, WSSV, IMNV, NHB, YHV. The last image illustrates the result of background elimination. The red circles indicate the disease expression.
6. Remarks
The experimentation has been conducted on shrimp images of 7 categories with a total of 348 samples. Removing the image’s background is the only step for the pre-processing procedure on collected images. The authors also tried the experiments without the step of background elimination, but the results were quite low. It is clearly stated in Table 4 that the combination of transfer learning and several convolutional neural networks works well in the shrimp images classification task. The inception-v3 model achieves the best classification accuracy score at 90.02%.
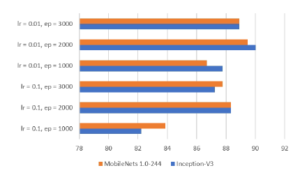
Figure 12: Accuracy comparison between two models.
One thing to note is that the shrimp’ images themselves have not appeared in the ImageNet dataset, on which the models have been pre-trained. The experiment results have proved the remarkable ability of knowledge transferability even if the dataset distribution is different. The best score of each version of the models is reached when the learning rate is 0.01, and the number of epoch is 2000.
By the time of conducting this paper, there are newer versions of inception and mobilenets. However, the authors choose inception-v3 and mobilenets-v1 because they are the standard transfer learning models, successfully integrated into many machine learning libraries, and can be adequately trained on a wide range of machines such as regular laptops, Google cloud TPU
(https://cloud.google.com/tpu/docs/inception-v3-advanced), smartphones, and small IoT devices (https://software.intel.com/enus/articles/inception-v3-deep-convolutional-architecture-forclassifying-acute-myeloidlymphoblastic). Newer versions need to be configured in different working environments.
7. Conclusion and Outlook
|
Table 4: The performance of two models regarding hyperparameters combination. The best scores are in bold.
|
|||||||||||||||||||||||||||||||||||||||||||
The growth of the shrimp industry is essential to provide shrimpbased products for global population demands, and the need for such manageable growth is intensively recognized as a principal goal. In the shrimp industry of Mekong delta of Vietnam, the farming sector is highly fragmented over regions. Shrimp farmers are vulnerable to epidemic diseases once they occur. Although various sources and agencies exist which aim to support services and treatments, there is not a single place where shrimp experts, farmers, computer scientists, treatment agencies, and policymakers can effectively connect. Shrimp farmers have to rely on neighbors and social networks for help on emerging diseases. By looking at the problems’ insight, the authors, throughout this work, pave several first stones of a complete solution. We have launched a data collection center that shrimp farmers assure to send images and questions. We apply the transfer learning technique to leverage the effectiveness of trained deep convolutional neural networks on different non-standard datasets. A good accuracy score is archived, confirming this research direction is worth investigating. We have investigated six common reported shrimp diseases and one class of healthy shrimp.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgment
This study was funded in part by the Can Tho University – Improvement Project VN14-P6, supported by a Japanese ODA. We thank our colleagues from the College of Aquaculture and Fisheries for advising on the fishery knowledge. The authors thank the farmers and anonymous contributors who participated and sent images used in the experiments. The discussions expressed are those of the authors and not necessarily those of the funding agency.
- N. T. P. Lan, “Social and ecological challenges of market-oriented shrimp farming in Vietnam,” SpringerPlus, 2(1), 1–10, 2013, doi:10.1186/ 2193-1801-2-675.
- T. B. T. Nguyen, “Good aquaculture practices (VietGAP) and sustainable aquaculture development in Viet Nam,” in Resource Enhancement and Sus- tainable Aquaculture Practices in Southeast Asia: Challenges in Responsible Production of Aquatic Species: Proceedings of the International Workshop on Resource Enhancement and Sustainable Aquaculture Practices in Southeast Asia 2014 (RESA), 85–92, Aquaculture Department, Southeast Asian Fisheries Development Center, 2015.
- A. Shinn, J. Pratoomyot, D. Griffiths, T. Trong, N. Vu, P. Jiravanichpaisal, M. Briggs, “Asian shrimp production and the economic costs of disease,” Asian Fish. Sci. S, 31, 29–58, 2018, doi:10.33997/j.afs.2018.31.S1.003.
- V. Boonyawiwat, N. T. V. Nga, M. G. Bondadreantoso, “Risk Factors Asso- ciated with Acute Hepatopancreatic Necrosis Disease (AHPND) Outbreak in the Mekong Delta, Viet Nam,” Asian Fisheries Society, 31, 226–241, 2018, doi:10.33997/j.afs.2018.31.S1.016.
- S. Thitamadee, A. Prachumwat, J. Srisala, P. Jaroenlak, P. V. Salachan, K. Sritunyalucksana, T. W. Flegel, O. Itsathitphaisarn, “Review of current disease threats for cultivated penaeid shrimp in Asia,” Aquaculture, 452, 69– 87, 2016, doi:10.1016/j.aquaculture.2015.10.028.
- R. W. Doyle, “Inbreeding and disease in tropical shrimp aquaculture: a reappraisal and caution,” Aquaculture research, 47(1), 21–35, 2016, doi: 10.1111/are.12472.
- G. Braun, M. Braun, J. Kruse, W. Amelung, F. G. Renaud, C. M. Khoi, M. Duong, Z. Sebesvari, “Pesticides and antibiotics in permanent rice, al- ternating rice-shrimp and permanent shrimp systems of the coastal Mekong Delta, Vietnam,” Environment international, 127, 442–451, 2019, doi:10.1016/ j.envint.2019.03.038.
- B. K. Dey, G. H. Dugassa, S. M. Hinzano, P. Bossier, “Causative agent, diag- nosis and management of white spot disease in shrimp: A review,” Reviews in Aquaculture, 2019, doi:10.1111/raq.12352.
- T. J. Sullivan, A. K. Dhar, R. Cruz-Flores, A. G. Bodnar, “Rapid, CRISPR- Based, Field-Deployable Detection Of White Spot Syndrome Virus In Shrimp,” Scientific Reports, 9(1), 1–7, 2019, doi:10.1038/s41598-019-56170-y.
- J. Xiong, W. Dai, C. Li, “Advances, challenges, and directions in shrimp disease control: the guidelines from an ecological perspective,” Applied microbiology and biotechnology, 100(16), 6947–6954, 2016, doi:10.1007/ s00253-016-7679-1.
- T. Q. Bao, T. C. Cuong, N. D. Tu, L. T. Hieu, et al., “Designing the Yellow Head Virus Syndrome Recognition Application for Shrimp on an Embedded System,” Exchanges: The Interdisciplinary Research Journal, 6(2), 48–63, 2019, doi:10.31273/eirj.v6i2.309.
- Z. Liu, F. Cheng, W. Zhang, “Identification of soft shell shrimp based on deep learning,” in 2016 ASABE Annual International Meeting, 1, American Society of Agricultural and Biological Engineers, 2016, doi:10.13031/aim. 20162455470.
- M. Ghasemi-Varnamkhasti, R. Goli, M. Forina, S. S. Mohtasebi, S. Shafiee, M. Naderi-Boldaji, “Application of image analysis combined with computa- tional expert approaches for shrimp freshness evaluation,” International Journal of Food Properties, 19(10), 2202–2222, 2016, doi:10.1080/10942912.2015. 1118386.
- H. Lu, Y. Li, T. Uemura, Z. Ge, X. Xu, L. He, S. Serikawa, H. Kim, “FD- CNet: filtering deep convolutional network for marine organism classifica- tion,” Multimedia tools and applications, 77(17), 21847–21860, 2018, doi: 10.1007/s11042-017-4585-1.
- Z. Liu, X. Jia, X. Xu, “Study of shrimp recognition methods using smart networks,” Computers and Electronics in Agriculture, 165, 104926, 2019, doi:10.1016/j.compag.2019.104926.
- Y. LeCun, L. Bottou, Y. Bengio, P. Haffner, “Gradient-based learning applied to document recognition,” Proceedings of the IEEE, 86(11), 2278–2324, 1998, doi:10.1109/5.726791.
- Z. Liu, “Soft-shell Shrimp Recognition Based on an Improved AlexNet for Quality Evaluations,” Journal of Food Engineering, 266, 109698, 2020, doi: 10.1016/j.jfoodeng.2019.109698.
- L. Torrey, J. Shavlik, “Transfer learning,” in Handbook of Research on Machine Learning Applications and Trends: Algorithms, Methods, and Techniques, 242– 264, IGI Global, 2010, doi:10.4018/978-1-60566-766-9.ch011.
- K. Weiss, T. M. Khoshgoftaar, D. Wang, “A survey of transfer learning,” Journal of Big Data, 3(1), 9, 2016, doi:10.1186/s40537-016-0043-6.
- M. Christopher, A. Belghith, C. Bowd, J. A. Proudfoot, M. H. Goldbaum, R. N. Weinreb, C. A. Girkin, J. M. Liebmann, L. M. Zangwill, “Performance of Deep Learning Architectures and Transfer Learning for Detecting Glaucomatous Optic Neuropathy in Fundus Photographs,” Scientific reports, 8(1), 16685, 2018, doi:10.1038/s41598-018-35044-9.
- N. Duong-Trung, L.-D. Quach, C.-N. Nguyen, “Learning Deep Transfer- ability for Several Agricultural Classification Problems,” International Jour- nal of Advanced Computer Science and Applications, 10(1), 2019, doi: 10.14569/IJACSA.2019.0100107.
- N. Duong-Trung, L.-D. Quach, M.-H. Nguyen, C.-N. Nguyen, “Classifica- tion of Grain Discoloration via Transfer Learning and Convolutional Neural Networks,” in Proceedings of the 3rd International Conference on Machine Learning and Soft Computing, 2732, Association for Computing Machinery, New York, NY, USA, 2019, doi:10.1145 0997.
- N. Duong-Trung, L.-D. Quach, M.-H. Nguyen, C.-N. Nguyen, “A Combination of Transfer Learning and Deep Learning for Medicinal Plant Classification,” in Proceedings of the 2019 4th International Conference on Intelligent Informa- tion Technology, 8390, Association for Computing Machinery, New York, NY, USA, 2019, doi:10.1145/3321454.3321464.
- J. Deng, W. Dong, R. Socher, L.-J. Li, K. Li, L. Fei-Fei, “Imagenet: A large- scale hierarchical image database,” 2009, doi:10.1109/CVPR.2009.5206848.
- O. Russakovsky, J. Deng, H. Su, J. Krause, S. Satheesh, S. Ma, Z. Huang, Karpathy, A. Khosla, M. Bernstein, et al., “Imagenet large scale visual recog- nition challenge,” International journal of computer vision, 115(3), 211–252, 2015, doi:10.1007/s11263-015-0816-y.
- M. Everingham, L. Van Gool, C. K. Williams, J. Winn, A. Zisserman, “The pascal visual object classes (voc) challenge,” International journal of computer vision, 88(2), 303–338, 2010, doi:10.1007/s11263-009-0275-4.
- J. Yosinski, J. Clune, Y. Bengio, H. Lipson, “How Transferable Are Features in Deep Neural Networks?” in Proceedings of the 27th International Conference on Neural Information Processing Systems – Volume 2, NIPS14, 33203328, MIT Press, Cambridge, MA, USA, 2014, doi:10.5555/2969033.2969197.
- O. Russakovsky, J. Deng, Z. Huang, A. C. Berg, L. Fei-Fei, “Detecting avoca- dos to zucchinis: what have we done, and where are we going?” in Proceedings of the IEEE International Conference on Computer Vision, 2064–2071, 2013, doi:10.1109/ICCV.2013.258.
- M. Oquab, L. Bottou, I. Laptev, J. Sivic, “Learning and transferring mid-level image representations using convolutional neural networks,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 1717–1724, 2014, doi:10.1109/CVPR.2014.222.
- H.-C. Shin, H. R. Roth, M. Gao, L. Lu, Z. Xu, I. Nogues, J. Yao, D. Mol- lura, R. M. Summers, “Deep convolutional neural networks for computer- aided detection: CNN architectures, dataset characteristics and transfer learn- ing,” IEEE transactions on medical imaging, 35(5), 1285–1298, 2016, doi: 10.1109/TMI.2016.2528162.
- C. Szegedy, W. Liu, Y. Jia, P. Sermanet, S. Reed, D. Anguelov, D. Erhan, V. Vanhoucke, A. Rabinovich, “Going deeper with convolutions,” in Proceed- ings of the IEEE conference on computer vision and pattern recognition, 1–9, 2015, doi:10.1109/CVPR.2015.7298594.
- S. Ioffe, C. Szegedy, “Batch normalization: Accelerating deep network training by reducing internal covariate shift,” arXiv preprint arXiv:1502.03167, 2015.
- C. Szegedy, V. Vanhoucke, S. Ioffe, J. Shlens, Z. Wojna, “Rethinking the inception architecture for computer vision,” in Proceedings of the IEEE con- ference on computer vision and pattern recognition, 2818–2826, 2016, doi: 10.1109/CVPR.2016.308.
- A. G. Howard, M. Zhu, B. Chen, D. Kalenichenko, W. Wang, T. Weyand, Andreetto, H. Adam, “Mobilenets: Efficient convolutional neural networks for mobile vision applications,” arXiv preprint arXiv:1704.04861, 2017.
- P. Srisapoome, K. Hamano, I. Tsutsui, K. Iiyama, “Immunostimulation and yellow head virus (YHV) disease resistance induced by a lignin-based pulping by-product in black tiger shrimp (Penaeus monodon Linn.),” Fish & shellfish immunology, 72, 494–501, 2018, doi:10.1016/j.fsi.2017.11.037.
- M. E. Frischer, R. F. Lee, A. R. Price, T. L. Walters, M. A. Bassette, R. Verdiyev, C. Torris, K. Bulski, P. J. Geer, S. A. Powell, et al., “Causes, diagnostics, and distribution of an ongoing penaeid shrimp black gill epidemic in the US South Atlantic Bight,” Journal of Shellfish Research, 36(2), 487–500, 2017, doi:10.2983/035.036.0220.
- S. Senapati, G. P. Kumar, C. B. Singh, K. M. Xavier, M. Chouksey, B. Nayak,
A. K. Balange, “Melanosis and quality attributes of chill stored farm raised whiteleg shrimp (Litopenaeus vannamei),” Journal of Applied and Natural Science, 9(1), 626–631, 2017, doi:10.31018/jans.v9i1.1242. - B. Verbruggen, L. K. Bickley, R. Van Aerle, K. S. Bateman, G. D. Stentiford, E. M. Santos, C. R. Tyler, “Molecular mechanisms of white spot syndrome virus infection and perspectives on treatments,” Viruses, 8(1), 23, 2016, doi: 10.3390/v8010023.
- K. P. Prasad, K. Shyam, H. Banu, K. Jeena, R. Krishnan, “Infectious My- onecrosis Virus (IMNV)–An alarming viral pathogen to Penaeid shrimps,” Aquaculture, 477, 99–105, 2017, doi:10.1016/j.aquaculture.2016.12.021.
- J. M. Leyva, M. Martinez-Porchas, J. Hernandez-Lopez, F. Vargas-Albores, T. Gollas-Galvan, “Identifying the causal agent of necrotizing hepatopancreati- tis in shrimp: Multilocus sequence analysis approach,” Aquaculture Research, 49(5), 1795–1802, 2018, doi:10.1111

