A Novel Representative k-NN Sampling-based Clustering Approach for an Effective Dimensionality Reduction-based Visualization of Dynamic Data
Volume 5, Issue 4, Page No 08-23, 2020
Author’s Name: Dharamsotu Bheekyaa), Kanakapodi Swarupa Rani, Salman Abdul Moiz, Chillarige Raghavendra Rao
View Affiliations
School of Computer and Information Sciences, University of Hyderabad, Hyderabad, Telangana, 500046, India.
a)Author to whom correspondence should be addressed. E-mail: 15mcpc21@uohyd.ac.in
Adv. Sci. Technol. Eng. Syst. J. 5(4), 08-23 (2020); ![]() DOI: 10.25046/aj050402
DOI: 10.25046/aj050402
Keywords: Dimensionality Reduction, Exploratory Analysis, Visualization, Clustering, Sampling, t-Distributed Stochastic Neighbor, Embedding, Cluster Validation Index, Interpolation
Export Citations
Visualization plays a crucial role in the exploratory analysis of Big Data. The direct visualization of Big Data is a challenging task and difficult to analyze. Dimensionality Reduction techniques extract the features in the context of visualization. Due to the unsupervised and non-parametric nature, most of the dimensionality reduction techniques are not evaluated quantitatively and not allowed to extend for dynamic data. The proposed representative k-NN sampling-based clustering, determines the underlying structure of the data by using well-known clustering techniques. The external cluster validation index determines the order sequence of clustering techniques from which the appropriate cluster techniques are recommended for the given datasets. From the recommended set, the samples of the best clustering technique are considered as representative samples which can be used for gener- ating the visual representation. The t-Distributed Stochastic Neighbor Embedding (t-SNE) algorithm is applied to generate a low-dimensional embedding model of representative samples, which is more suitable for visualization. The new data samples are added to the generated model by using the interpolation technique. The low-dimensional embedding results are quantitatively evaluated by k-NN accuracy and trustworthiness. The performance analysis of representative k-NN sampling-based clustering results and embedding results accomplished by seven di datasets.
Received: 13 May 2020, Accepted: 19 June 2020, Published Online: 06 July 2020
1. Introduction
Exploratory analysis of Big Data is ubiquitous in an increasing number of fields and vital to their progress. Visualization plays a paramount role in an exploratory study. Data visualization is applicable for the limited number of dimensions, which depends on the perceptual capability of the analyst. For exploratory analysis, traditional visualization techniques may not provide useful visual insights of high-dimensional data and they are restricted for a limited number of dimensions. The conventional feature selection methods may not provide helpful visual insights for exploratory analysis, which happens due to the inappropriate feature selection. There is a requirement of the feature extraction technique, which shows the correlation between the original features of the data. The Dimensionality Reduction (DR) technique transforms the data and extract new features, which makes data analysis tractable. From the last few decades, researchers have proposed various linear as well as non-linear DR techniques in the context of visualization. The linear DR techniques like Principal Component Analysis (PCA) [1], Multidimensional Scaling (MDS) [2] and Factor Analysis (FA) [3] In contradiction to linear DR, the non-linear DR techniques like Isomap [4], Local Linear Embedding (LLE) [5], Laplacian Eigenmap (LE) [6] and Stochastic Neighbor Embedding (SNE) [7] deals with non-linear data. The paper [8] provides a complete comparative study of DR techniques. The methodology of the DR technique depends on its feature extraction criteria. The feature extraction depends on the characteristics of interest in the data such as interpoint distances, reconstruction weights, variation, linear subspace, geodesic distances, linear tangent space, neighborhood graph and conditional probability distribution.
Among all the DR techniques, t-Distributed Stochastic Neighbor Embedding (t-SNE) [9], is an improved version of SNE, introduced by Laurens Van Der Maaten and Geoffrey Hinton in 2008. The t-SNE has gained impressive attention and enormous popularity in several fields [10–12]. The empirical study states that the lowdimensional visual representation of t-SNE is more robust than any other DR technique. The t-SNE algorithm most commonly used to preserve the original structure of the high-dimensional data in very low-dimensional embedding (i.e., either 2D or 3D). From past several years the t-SNE has been explored in various aspects such as optimization [13,14], scalability [14–17], dealing with non-numeric data [18], outliers separation [17] and many more.
The t-SNE is a non-parametric technique that provides flexibility in learning and reduces computational complexity. The nonparametric nature limits t-SNE applicability to the out-off-sample extension, which means the addition of new data samples into the existing t-SNE environment is not possible. If we want to add a new sample, then we should re-run the entire t-SNE model by including a new sample. When the addition of new data points increases, the computational cost of t-SNE also increases monotonically. Therefore, it does not apply to time-series and streaming data. The LION-tSNE [17] of Boytsov et al. addressed the problem of adding new data into the existing t-SNE environment using Local Inverse Distance Weighting Interpolation (LIDWI) without re-running. The outlier’s handing is also addressed by LION-tSNE using outlier placement heuristic, which assumes that some percentile of outliers present in the designed t-SNE and determine the outliers from the newly added data points. In LION-tSNE, the sample t-SNE model is designed based on the random sample selection, which may cause the non representativeness of the data. The representative samples are selected by our earlier approach called k-NN sampling [19] and the resutlts are statistically significant which is measured by statistical method pairwise t-test [20].
This paper is an extension of our earlier paper presented in High Performance Computing, Data and Analytics (HiPC) [19], which deals with the preservation of the underlying cluster structure of high-dimensional data in low-dimensional t-SNE embedding with a representative sample. The underlying cluster structure preservation is measured in terms of a quantitative metric. In the existing methods, the low-dimensional embedding of t-SNE describes the quality of the structure preservation. Still, it is an open problem for giving the quantitative proof for the number of clusters that exist in the original data. The proposed novel representative k-NN sampling-based clustering approach for effective dimensionality reduction-based visualization finds the solution. In the first step, the proposed approach determines several distinct data samples using our earlier proposed k-NN sampling algorithm. The number of samples depends on the range of k (i.e., 1 ≤ k ≤ m), which gives the neighborhood representation.
In the second step, the effective number of clusters existing in the original dataset is determined by the sampled data using clustering techniques such as k-Means [21], Agglomerative Hierarchical Clustering (AHC) [22], Balanced Iterative Reducing and Clustering using Hierarchies (BIRCH) [23], Fuzzy c-means (FCM) [24], Density-Based Spatial Clustering of Applications with Noise (DBSCAN) [25], Ordering Points To Identify the Clustering Structure (OPTICS) [26], Mean Shift (MS) [27], Spectral Clustering (SC) [28], Expectation-Maximization Gaussian Mixture Model (EMGMM) [29], Affinity Propagation (AP) [30] and Mini Batch k-Means (MBKM) [31]. Each clustering technique generates cluster labels for sampled data. The cluster labels of remaining data (i.e., data points other than selected samples) are labeled by the k-Nearest Neighbor (k-NN) algorithm. The k-NN of each remaining data sample is subject to the selected samples. After assigning the labels to the remaining data samples, the representative sample of each clustering technique is determined by their external cluster validation index called Fowlkes-Mallows Index (FMI) [32]. In our contribution, we are also recommending the order sequence of clustering techniques among the selected techniques for a given dataset. We are also presenting the Compactness (CP) [33], Calinski-Harabaz Index (CHI) [33] and Contingency Matrix (CM) [34] of clustering techniques for gaining a more detailed analysis about clustering. The first technique in the order sequence denotes the best clustering technique and the representative samples of it considered as the samples for t-SNE model design for a given dataset. The cluster validation index provides the comparison between representative k-NN sampling-based clustering and the aggregate clustering (i.e., clustering on the whole dataset). The proposed representative k-NN sampling-based clustering is scalable to all clustering techniques which are suitable for numerical datasets. Also, we can apply any clustering technique to the large scale dynamic data with representative k-NN sampling-based clustering. Due to the paper limitation, we have chosen the most popular clustering techniques from different groups.
In the third step, the sample t-SNE model is designed on a representative sample of best clustering techniques, which transforms the high-dimensional data into low-dimensional embedding. The remaining data samples are added to the sample t-SNE environment with the help of LIDWI. The outliers from the remaining samples are identified and controlled by proposed heuristic and the identified outliers are placed into the t-SNE environment using outlier placement heuristic of Boystov et.al [17]. In the fourth step, the t-SNE embedding of input data is quantitatively evaluated by k-NN accuracy in the context of clustering and trustworthiness. The quantitative evaluation answers the question, how much structure of high-dimensional data is preserved by the low-dimensional t-SNE embedding. The k-NN accuracy of t-SNE embedding is measured in two ways: baseline accuracy and sampling accuracy. For quantitative performance evaluation, the k-NN accuracy of t-SNE embedding of representative k-NN sampling-based clustering and aggregate clustering results are compared with the ground truth class labels, which is measured in our earlier paper. In our earlier approach, we used ground-truth class labels for obtaining the k-NN accuracy of the t-SNE embedding but here we are using for checking the derived cluster purity of representative k-NN sampling-based clustering. The k-NN sampling-based clustering results and t-SNE embedding of it are analyzed by seven differently characterized toy and real-world datasets. In summary, our contribution consists of
- The order sequence of applicable clustering methods with representative k-NN sampling-based clustering using cluster validation index. The set of recommended techniques for the given dataset using a threshold. The comparison between representative k-NN sampling-based clustering and aggregate clustering results.
- The outliers from the remaining samples are identified and controlled by the proposed heuristic before adding into the t-SNE embedding. The embedding structure quantitatively evaluated by k-NN accuracy in the context of clustering and trustworthiness.
The organization of the paper is as follows. In section 2, we present the background knowledge that gives the basic mathematical intuition required for understanding the clustering techniques, t-SNE and other supplements. Section 3 presents the related work. In section 4, we present a detailed description of the proposed k-NN sampling-based clustering for the visualization of dynamic data. In section 5, we are providing the details of datasets, experimental setup and analysis of results. Finally, section 6 gives directions for future work and conclusions.
2. Background Knowledge
The following section describes the few techniques for understanding the background formulation of our work. Section 2.1 gives a detailed description of the selected clustering techniques for representative k-NN sampling-based clustering. Section 2.2 describes the mathematical intuition of the t-SNE algorithm. The intuition behind the addition of the inlier data point into an existing t-SNE environment is explained in section 2.3. In section 2.4, we are giving a detailed description of metrics that are used for the quantitative evaluation of embedding.
2.1 Clustering and their performance measures
Section 2.1.1 describes the selected clustering techniques which are used for representative k-NN sampling-based clustering. 2.1.2 provides detailed mathematical intuition of cluster validation indexes such as Fowlkes-Mallows Index (FMI), Compactness (CP), Calinski-Harabaz Index (CHI) and Contingency Matrix (CM).
2.1.1 Clustering Techniques
Clustering is unsupervised learning, where the similarly characterized data objects are grouped. The clusters of data objects can be represented as a set C of subsets C1,C2,….,Ck such that . The different clustering algorithms are proposed based on their measures of similarity: partitional, hierarchical, fuzzy theory-based, distribution-based, density-based, graph partition-based, grid-based, model-based and many more. The user decides the number of clusters present in the dataset by using a heuristic, trail-and-error and evolutionary approaches such as density and probability density. From the groups mentioned above, we have chosen the most frequently and popularly used algorithms for experimental evaluation. But proposed representative k-NN sampling-based clustering approach is scalable to all the clustering techniques which are suitable for the numerical datasets. We explored traditional clustering algorithm [35–37] for our work.
In partitional clustering, data points assigned to any one of the k-clusters using distance similarity measures such as Euclidean distance. k-means [21] clustering is one of the simplest, best-known and benchmarked partition-based clustering. The k-means clustering classifies the given data points through a user-defined number of clusters. The main goal of the k-means clustering is the initialization of an appropriate k-centroids, one for each cluster. The objective of k-means is the minimization of the sum of square distance which can be defined as follows
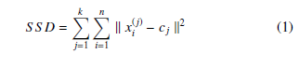
where k xi(j) − cj k2 is a L2 norm between a data point xi(j) and the cluster center cj. In AP [30], at the initial stage, all data points are considered as centroids and nodes of the network. The clusters and their centroids are measured by transmitting the similarity message recursively. The Mini Batch k-Means (MBKM) [31] is an improved version of k-means, which performs clustering on batches instead of considering each point. Therefore, MBKM requires less computational time and is also applicable to large datasets.
In hierarchical clustering, groups are formed by iteratively dividing the data objects either in the bottom-up or top-down approach. The bottom-up approach is also known as Agglomerative Hierarchical Clustering (AHC) [22], in which initially each data object is considered as a separate group, then merging these small groups into larger and larger groups until a single group formation or certain threshold. The top-down approach is also known as Divisive Hierarchical Clustering (DHC), and it works in reverse order of the bottom-up approach. Both hierarchical approaches depend on the linkage criteria such as single, complete and average linkage. The linkage criteria determines the metric for merging two similar characterized small clusters. For example, single linkage criteria determines the minimum distance pair from the neighbor clusters minnd(x,y) : x ∈ Ci,y ∈ Cjo. The BIRCH [23] method has proposed to deal with large datasets, outliers in robust and also to reduce the computational complexity. The BIRCH method works on the idea of Cluster Features (CF) which is a height-balanced tree.
The basic idea of fuzzy theory-based clustering is that the discrete labeling is converted to continuous intervals, to describe the belonging relationship among objects more reasonably. The Fuzzy c-means (FCM) [24] is an extension of k-means where each data point can be a member of multiple clusters with membership value. The main advantage of FCM is that the formed groups are more realistic.
The density-based clustering finds the clusters based on the density of data points in a region. The principal objective of densitybased clustering is that for each instance of a group, there should be at least a minimum number of neighbor instances within the given radius. The DBSCAN [25] is the most well known density-based clustering. In DBSCAN clustering, the data objects fall into three groups: core-object, border-object and noise-object. The data points of the core-object group have enough number of neighbors in the given radius, and these data points are from the higher density region. The data points of the border-object group have fewer neighbors than the required number of neighbors in the given radius and they are in the neighborhood of the core object group. The data points of the noise-object group are not in either core or border-object group. The advantage of DBSCAN clustering is that the generated clusters are in arbitrary shape based on the given parameters such as radius and number of minimum instances. The improved version of DBSCAN is OPTICS [26], which overcomes the limitations of DBSCAN. Mean-shift [27] algorithm determines the mean of offset of the current data point, the next point is identified based on the current point and the offset. The process will continue until some criteria is satisfied.
The spectral clustering [28] is a graph partition-based technique in which clustering is obtained by similarity graph partition. The paper [28] describes the different ways of constructing the similarity graph. While building the similarity graph, the data objects are considered as vertices and the similarity (i.e., the similarity distance) between the data objects is a weighted edge. The critical idea of similarity measure is to find a method of graph partition by making edges between different groups have low edge weight and the edges of the same group have more edge weights. In similarity graph partitioning, the cluster analysis is carried out by feature space obtained by the eigenvectors corresponding to k largest eigenvalues of the laplacian matrix [38] of similarity graph. The spectral clustering is suitable for the datasets with arbitrary shape, high-dimensions and outliers.
The EMGMM [29] is a model as well as a distribution-based clustering technique in which cluster data obey the same independent Gaussian distribution which exists in the original data distribution. The generated clusters are more realistic because a probability distribution obtains them.
The performance and applicability of clustering depends on the used dataset characteristics. Therefore, the researchers empirically state that none of the clustering technique is generalized for any datasets.
2.1.2 Cluster validation Measures
In our experimentation, we have used two internal and one external cluster validation measure to evaluate the performance of clustering techniques. The internal measure computes the performance of clustering without using the ground-truth class labels. The compactness (CP) [33] is an internal cluster validation measure which can be computed as follows
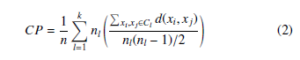
where d(xi, xj) is Euclidean distance between two objects in cluster Cl and nl is the number of objects in Cl. The smaller CP value of clustering is more compact and gives better clustering results. The Calinski-Harabaz Index (CHI) [33] or Variance Ratio Criterion is defined as the ratio of between-clusters dispersion mean and the within-cluster dispersion mean. The CHI is an internal cluster validation measure which is given by
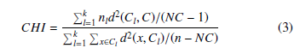
Where n is the dataset size, C is the dataset center, nl is the size of lth cluster, Cl is the lth cluster centroid and NC is the number of clusters.
The external validation measure knows the ground truth class labels. The primary purpose of the external validation index is to choose an optimal clustering algorithm for a given dataset. The external validation measure also checks the cluster purity. In our experiment, we used Fowlkes-Mallows Index (FMI) [32] is the geometric mean between precision and recall. The FMI defined as follows
![]()
The FMI value is bounded between 0 and 1, 1 denotes that the obtained clusters and the given ground truth classes are the same. The large FMI indicates that the obtained clustering is purer.
We also measured the contingency matrix that reports the intersection cardinality for every true/predicted cluster pair. The contingency matrix gives sufficient statistics for all clustering metrics. But it is hard to interpret the contingency matrix of extensive clustered data.
2.2 Student t-Distributed Stochastic Neighbor Embedding (t-SNE)
The t-SNE [9] algorithm introduced by Laurens Van Der Maaten and Geoffrey Hinton in 2008, based on the SNE algorithm. The principal objective of SNE is to preserve the underlying structure of high-dimensional data in low-dimensional embedding space. Lets assume the given input data X = {x1, x2,….., xN} where each xi ∈ RD is a D-dimensional vector. The t-SNE computes the embedding Y = {y1,y2,….,yN} of X where each yi ∈ Rd is a d-dimensional vector, where d D and most commonly d =2 or 3. The similarity between xi and xj of X is calculated by conditional probability pij is given by
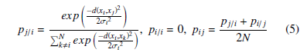
where the bandwidth σi of Gaussian kernel, is obtained by binary search by matching the perplexity of Pi and the user-defined perplexity (µ) which is given as a parameter. The perplexity is a smooth measure of an adequate number of neighbors for each data point. The equality and perplexity of Pi is as follows
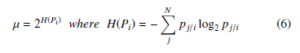
where H(Pi) denotes the entropy and Pi is the conditional probability distribution across all data points for the given xi. The yi and yj are the corresponding low-dimensional values of xi and xj (i.e., the values of Y are initialized by Gaussian or uniform distribution).
The similarity between yi and yj is defined as
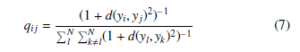
where d(yi,yj) is defined as distance similarity measure such as Euclidean distance. In low-dimensional embedding, the similarity qij is obtained by student t-distribution with one degree of freedom. But, the similarity pij of high-dimensional data uses the Gaussian distribution. The cumulative function curvature of Gaussian distribution is flatter than the cumulative function curvature of student t-distribution with one degree of freedom. The principal idea of using student t-distribution in low-dimensional embedding is to overcome the crowding problem [7].
If pij ∼ qij, ∀i,j ∈ N, then the given data is perfectly embedded into the low-dimensional space. Otherwise, compute the KL-divergence (i.e., error) between pij and qij that is equal to the cross-entropy in Information Retrieval System (IRS). The cost function (C) or objective of t-SNE is defined as follows
![]()
The simple gradient descent applied on cost function C for obtaining the optimization or minimization of it. The simple gradient descent of cost function C given by
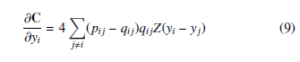
where Z = (1 + d(yi,yj)2)−1 is a normalization term of student tdistribution. The Equation 9 produce either positive or negative values depending on the pij value. If an Equation 9 gives positive value then there is an attractive force among the yi and yj of embedding space. Otherwise, there is a repulsive force among the yi and yj of embedding space. The degree of repulsion depends solely on the closeness of points in the embedding space. In the optimization process, the early exaggeration coefficient α > 1 plays a paramount role in forming groups of similar objects of high-dimensional data in low-dimensional embedding space. In the early exaggeration process, the elements of similarity matrix (P) (i.e., pij’s) multiplied by the early exaggeration coefficient, which is measured by the intuition given by George C. Linderman and Stefan Steinerberger [39]. Therefore, similar data objects bring near to each other in low-dimensional embedding space. This process can happen at the early stage of optimization. The gradient descent of C after early exaggeration is
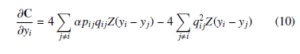
The momentum and learning rate parameters improve the optimization process of the cost function. The momentum parameter reduces the number of iterations of the cost function optimization. At the initial stage of iteration, the momentum value is small until the map points have become moderately well organized. The optimization is improved by input approximation and tree-based algorithms [13], which reduce the memory and computational complexity. The updated values of Y at iteration t is obtained by
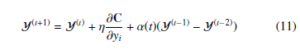
where α(t) is momentum at tth iteration, η is learning rate. The work-flow of t-SNE is shown in the Figure 1.
The well-separated clusters of input data are well preserved in the low-dimensional embedding by setting the early exaggeration coefficient α and learning rate η. The intuition for setting the above parameter is derived by George C. Linderman and Stefan Steinerberger in [39]. According to the George C. Linderman and Stefan Steinerberger observations the early exaggeration coefficient α, learning rate η, and minimum probability pij’s of same cluster objects(i.e., xi and xj belong into the cluster Cl for all i , j) is defined as follows.
![]()
where π : {1,2,….,n} → {1,2,…,k} assigns each data point to one of the k clusters.
2.3 Local Inverse Distance Weighting Interpolation (LIDWI)
The LIDWI [40] maps new data point x ∈ RD into the existing embedding yi ∈ Rd, where {i = 1,2,…….m}. The LIDWI determines the value of x as a weighted sum of values yi, where weight is proportional to inverse distances. The LIDWI of x is
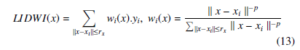
for instance, when the data point x → xi, the inverse distance k x − xi k−1→ ∞, the corresponding weight wi(x) → 1 ( i.e., ∀j,iwj(x) → 0 due to the normalization) and LIDWI(x) → yi. The neighbor points selection is obtained by a radius rx parameter. The parameter rx value is calculated by the heuristic proposed by Andrey Boytsov et.al. [17]. In LIDWI, the power parameter p plays an important role. For instance, very small value of p predicts the value of x around the center: y ≈ mean(yi) (unless x = xi) even the distance k x − xi k is low because the weight distribution is close to uniform. When the power parameter is high and the distance k x − xi k is low, the weight wi(x) of very first nearest neighbor is dominating all other neighbors, therefore y ≈ yi where i = argmin k x − xj k. The overfitting suffers from either too small or too large values of power parameter p. In LOIN-tSNE, the authors proposed a generalization for obtaining power parameter by using leave-one-out cross-validation of the training sample. The computation of the generalized power parameter is obtained by applying the LIDWI for each training sample that produces the estimation of each yi. Then the mean square distance between the estimated yi’s and real yi’s is computed. The optimal power parameter is obtained by optimizing the mean square error (i.e., the mean square distance is minimum). The obtained power parameter is considered as a metric. However, this metric is heuristic, not an exact criterion.
2.4 Performance evaluation metrics
2.4.1 k-NN accuracy
The existence of the cluster structure of high-dimensional data in low-dimensional embedding is quantitatively measured by k-NN accuracy of the t-SNE embedding in the context of clustering. The k-NN accuracy is defined as the percentage of the neighbors having the cluster label equivalent to the observational point cluster label.
2.4.2 Trustworthiness
Trustworthiness [41] is one of the measure to evaluate the quality of the t-SNE embedding. Trustworthiness is defined as any unexpected nearest neighbors in the output space are penalized in proportion to their rank in the input space.
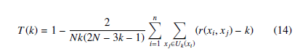
where Uk(xi) is the k-NN of xi in embedding space, r(xi, xj),i , j the rank of xj when the data vectors are ordered based on their Euclidean distance from the data vector xi in the original data space. It is bounded between 0 and 1, where 1 represents the complete structure of the data preserved in the embedding space.
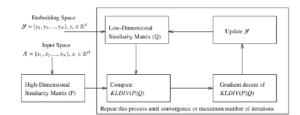
Figure 1: Work-flow of t-SNE algorithm [19]
3. Related Work
Our earlier proposed k-NN sampling based tSNE mainly concentrates on the preservation of the underlying structure of highdimensional data in low-dimensional embedding space with representative samples. The obtained low-dimensional embedding structure describes the quality of the structure using ground-truth class labels. But the low-dimensional embedding does not give the quantitative proof for the number of clusters that exist in the original data. In [39] authors gave the theoretical observations for well-separated clusters of high-dimensional data in low dimensional embedding space.
The researchers have proposed various methods to incorporate new data samples into the existing t-SNE environment. Most of the existing techniques designed a mapping function f : X → Y, which accepts multi-dimensional data and returns its low-dimensional embedding. The designed mapping functions are used for incorporating the new data samples into the existing t-SNE environment. In [14–16] authors have proposed different approaches for adding a new data sample or scaling up the t-SNE algorithm.
Andrey Boytsov et.al. [17] proposed the LION-tSNE algorithm based on local IDWI for adding new data sample into an existing tSNE environment. It also addresses the outlier handling approaches. Our earlier work extended the idea of the LION-tSNE algorithm by proposing a k-NN sampling method for designing a representative sampling based t-SNE model. It allows the selection of the sample concerning their k-nearest neighbors instead of random sampling. In this paper, we are proposing the novel representative k-NN sampling-based cluster approach for effective dimensionality reduction-based visualization of dynamic data, which determines the underlying cluster by using the most popular clustering techniques. The obtained cluster structure is quantitatively evaluated by k-NN accuracy in the context of clustering and trustworthiness.
4. Proposed representative k-NN samplingbased clustering for effective visualization Framework
The proposed representative k-NN sampling-based clustering for effective dimensionality reduction-based visualization of dynamic data framework is shown in the Figure 2. It has four phases: at phase 1, data preprocessing is done for removing the redundant data points and filling the empty variables with appropriate values. At phase 2, the proposed k-NN sampling-based clustering is applied to determine: (i) The generation of k distinct samples using mutual k-NN sampling with static graph updation algorithm. (ii) The list of clustering techniques applicable for the given dataset. (iii) The optimal sample size produces the best grouping for each concerned clustering technique. (iv) The ordered sequence of recommended clustering techniques for the given dataset from which the best clustering technique is selected by using FMI scores. The detailed representative k-NN sampling-based is clustering presented in section 4.1. At phase 3, the dimensionality reduction-based visualization is obtained by t-SNE algorithm, which derives the low-dimensional embedding of data and the LIDWI algorithm is used to interpolate the new data points into the learned t-SNE model. The outliers from the remaining data points are identified and controlled by the proposed heuristic. The dimensionality reduction-based visualization is described in section 4.2. In the final phase, the quantitative measure of the t-SNE embedding is computed by k-NN accuracy and trustworthiness. The process of quantitative metric derivation is described in section 4.3.
4.1 Representative k-NN sampling-based clustering
Initially, the k distinct samples are generated by the modified algorithm of our earlier proposed method. The sample generation depends on the parameter k of k-NN sampling. Each k generates one distinct sample; the boundary of k is denoted as 1 ≤ k ≤ m, where m is the maximum number of neighbors required for preserving the behaviour of any data point of the given dataset. Our earlier approach has given two different k-NN sampling strategies LION-tSNE with quantitative measure of cluster accuracy such as static and dynamic k-NN sampling based on k-NN graph modification. In this paper, we are considering only static k-NN sampling in which the k-NN graph is modified statically that means no new edge is added among any two vertices of the k-NN graph. The main reason for selecting static k-NN sampling is that the t-SNE embedding results of it are more statistically significant than the dynamic one. The Nearest Neighbor score (NN score) and Mutual Nearest Neighbor score (MNN score) of each node of the k-NN graph are used for selecting the samples. Lets assume k-NN graph is a directed graph G = (V, E), the edge E(v1,v2) gives v2 as a neighbor of v1 and neighborhood of v1 is denoted by Nv1. The out-degree of each vertex is equal to k, and the in-degree of a vertex depends on the neighborhood property of other vertices (i.e., the data point xi belong into the neighborhood of other datapoints). In our method, each data point is a vertex of the k-NN graph and k is a parameter for deriving the edges between neighboring vertices. The NN score of data point xi is equal to the in-degree of xi which is defined as follows
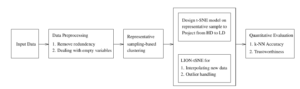
Figure 2: Proposed framework of representative k-NN sampling-based clustering for an effective dimensionality reduction-based visualization of dynamic data using

The updated mutual k-NN sampling with a static graph updation algorithm is shown in Algorithm 1. Initially, the Train samples (i.e., representative samples set) and Rep samples (i.e., the sample represented by the selected training sample that is the first nearest mutual neighbor of train sample) are null sets. For each iteration, the data point x which has maximum NN and maximum MNN score appended to the Train samples set. The data point y is appended to the Rep samples if y ∈ Nx and x ∈ Ny that denotes the point x and y are more similar (i.e., the distance d(x,y) < d(x,z) where ∀z ∈ Nx) to each other. The data point x and y are deleted from X and their corresponding vertices are deleted from the k-NN graph. After deleting the vertices of x and y, the corresponding in-edges and out-edges are removed from the k-NN graph and the graph is updated accordingly. The elements of Train samples and Rep samples are obtained iteratively, the iteration repeats until the NN score of the remaining X is equal to zero.
Algorithm 1: Mutual k-NN Sampling with a static graph updation
Data: data set X = {x1, x2,…..xN}, parameter k for minimal training sample selection
Result: Return Train samples, Rep samples
Train samples = ∅
In our earlier approach of static graph updation, we have considered the whole mutual neighborhood set of x as the samples represented by x. The whole neighborhood set selection causes loss of information due to that reason the earlier approach select Train samples set at an early stage of k value. The data points of the remaining X (i.e., the data point do not belong into either Train samples set or Rep samples set) is handled in two different ways. Handling of the remaining samples is described in subsection 4.4. The mutual k-NN sampling with a static graph updation algorithm generates a various set of Train samples and Rep samples with different k-values.
The clustering techniques which are described in subsection 2.1.1 are applied on each Train samples set of mutual k-NN sampling with a static graph updation algorithm that is called a samplingbased clustering. The cluster labels for the data points other than the Train samples are assigned by using k-NN algorithm. For example, the dataset size is N, the Train samples size is n and then the unmarked sample size is N − n(i.e., the samples are not assigned with any labels). The k-NN of each unmarked sample is derived from the associated Train samples set. The label assignment of each unmarked sample depends on the labels of its k-NN, a label with maximum occurrence in k-NN that is assigned to a sample. From each clustering technique, the optimal Train samples set is obtained by the FMI score. The FMI is an external validation index that uses the ground truth class labels. Therefore, the number of clusters is defined as a constant that is equal to the number of ground truth classes. The FMI score of optimal Train samples of clustering techniques generates an order sequence of the clustering techniques. From the order sequence, we can recommend the most desirable techniques for a given dataset from the selected set of techniques. From this recommendation, the best suitable technique is selected and its optimal Train samples set is considered as the best representative sample. The threshold parameter is used to derive the recommended techniques, which is defined as the FMI score difference between two adjacent techniques of order sequence. The three cluster validation index such as FMI score, CHI score and CP of representative k-NN sampling-based clustering are compared with their aggregate clustering (i.e., clustering on whole dataset) validation index of chosen clustering techniques. The result comparison is discussed in subsection 5.3. The algorithm of representative k-NN sampling-based clustering is shown in Algorithm 2. The embedding of selected optimal Train samples and addition of other data samples into an existing t-SNE environment is described in the following section.
4.2 Dimensionality reduction-based visualization
The subsection 4.2.1 describes the low-dimensional embedding of a representative sample with t-SNE algorithm. Subsection 4.2.2 describes the addition of new data samples into an existing t-SNE environment that is called out-off-sample extension.
4.2.1 Low-dimensional embedding of a representative sample with t-SNE algorithm
Barnes-Hut t-SNE (BH-tSNE) [13] algorithm is an optimized version of the t-SNE algorithm. BH-tSNE optimizes the t-SNE objective function by input similarity approximation and gradient descent approximation. Therefore, it generates low-dimensional embedding of data with minimum computational and memory complexity than original t-SNE. In our approach, the BH-tSNE algorithm is used in two different ways for calculating the low-dimensional embedding space: 1. Baseline t-SNE embedding, 2. Sampled t-SNE embedding. The baseline t-SNE embedding is obtained by applying BH-tSNE on the whole dataset. In contrast, the sampled t-SNE embedding is obtained by applying BH-tSNE on the best representative sample, which is selected from the representative k-NN sampling-based clustering. The Baseline t-SNE embedding results analyze the overall structure of the data in low-dimensional embedding. The sampled t-SNE embedding results analyze the data structure with sampled data and allows the addition of new data samples into an existing t-SNE environment, which solves the scalability issue of the t-SNE. For obtaining a well-separated cluster in low-dimensional t-SNE embedding, the value of early exaggeration coefficient α, learning rate η and input similarity probability pij’s are adjusted according to the George C.linderman and Stefan Steinerberger intuition. In our experimentation, the initial solutions of t-SNE is assigned in three different ways, such as random, PCA based and MDS based initial solutions. The random initial solution takes many iterations for convergence. The PCA and MDS based initial solutions overcome the problem of random initialization and they produce better accuracy results, but their initial solution is cost-effective. Adding new data points into a designed t-SNE model is discussed in the below section.
4.2.2 Out-off-sample extension: Interpolation and Outlier handling
The addition of new data point to t-SNE embedding depends on the parameter rx, ry and rclose values. The value of parameter rx, ry and rclose is obtained from the best representative sample and the t-SNE embedding of it. The parameter rx is defined as the percentile of the 1-NN distance of a representative sample that decides whether the given new data point is either inlier or outlier. In our proposal, we came to know that the objects of the representative sample set are representing at least one sample of the dataset. Therefore, the representative sample set does not contain any outlier object and we have considered the parameter rx as the maximum 1NN distance of it. If the new data point x has at least one data point within the rx from the representative sample set, then x is an inlier, otherwise outlier. The dilation factor (df) is used to derive a heuristic rx = (1+d f) ∗ rx which controls the consideration of outliers percentage. The LIDWI interpolation technique is used for adding an inlier data point to the t-SNE embedding of the representative sample set. The outlier placement depends on the parameter ry and rclose. The outliers placed into the t-SNE embedding of the representative sample set according to the heuristic of Boytsov et.al. [17]. The k-NN accuracy and trustworthiness of t-SNE embedding quantitatively evaluate the existence of a high-dimensional cluster in the low-dimensional embedding. The quantitative evaluation described in the following section.
4.3 Quantitative metric derivation: k-NN accuracy
In our experimentation, the k-NN accuracy is calculated in two different ways: baseline accuracy and sampling accuracy. The kNN accuracy of t-SNE embedding of aggregate data is known as baseline accuracy. The sampling accuracy is computed in three different forms such as training accuracy, test accuracy and overall accuracy. The k-NN accuracy of t-SNE embedding of the representative sample is known as training accuracy. The k-NN accuracy of interpolated samples (i.e., the samples other than the representative sample) is known as test accuracy. The k-NN accuracy of combined low-dimensional space (i.e., the integration of t-SNE embedding of sampled data and interpolated data) is known as overall accuracy. The k-NN accuracy typically depends on the parameter k, which is considered as fixed in our experimentation which is discussed in subsection 5.2. For instance, the smaller k will give good performance accuracy and while increasing the k, performance accuracy will decrease. The selection of parameter k also plays a paramount role in k-NN accuracy measure. The relationship between the parameter k and accuracy is shown in the Figure 3. For quantitative performance evaluation, the k-NN accuracy of representative k-NN samplingbased clustering and aggregate clustering are compared with the k-NN accuracy of ground truth class labels, which is discussed in subsection 5.3.
4.4 t-SNE sample selection criteria
In our experimentation, we have considered two different sample selection criteria for designing the t-SNE model. In the first criteria, the samples are mostly representative of at least one or more other samples. The samples are obtained by a mutual k-NN sampling with a static graph updation algorithm and the best representative sample is selected by the representative k-NN sampling-based clustering. We observe that most of these samples were chosen from the dense region because they produce good NN score and MNN score.
Therefore, these samples do not contain any outliers. In this criterion, the outliers from the remaining sample addition are not sufficiently identified by the adequate 1-NN distance. The outliers consideration is controlled by the proposed heuristic, which is discussed in subsection 4.2.2. In the second criteria, the data points added to the representative sample from the remaining data points using (t,m, s)-Nets sampling [42]. For the addition of (t,m, s)-Nets samples, we used the same procedure of our earlier work. The (t,m, s)-Nets select samples randomly in a uniform distribution. These samples may not represent any other samples and the samples may change among the executions due to the randomness. Therefore, the samples of this criteria may contain outliers. The outliers of this criteria are handled in similar ways as they are handled in our earlier approach.
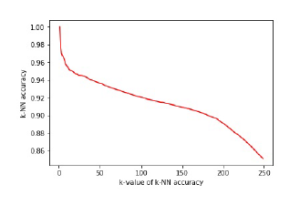
Figure 3: Relationship between k-value of k-NN accuracy and k-NN accuracy which is obtained from the baseline accuracy of k-NN sampling-based clustering.
5. Experimental result analysis
Here we are briefing the datasets, experimental setup and the result analysis. Section 5.1 describes the numerical datasets of different characterizations. The details about the experimental setup is given in section 5.2. The results analysis of each dataset is described in section 5.3.
5.1 Datasets
In our experimental evaluation, we have considered differently characterized datasets such as IRIS, Breast-Cancer, Leukemia, Wine, MNIST, Olivetti-Faces, and COIL-20. The Table 1 provides a detailed description of all the datasets which are downloaded from the UCI Machine Learning repository [43]. For our experimentation, the features of IRIS, Breast-Cancer and Wine datasets are normalized between zero and one, which improves the computational complexity. Initially, the KernelPCA [44] is applied to reduce the dimensionality of the high-dimensional datasets such as Leukemia, MNIST, Olivetti-Faces and COIL-20. The Leukemia dataset is a micro-array of gene expression. The MNIST, Olivetti-Faces and COIL-20 are image datasets that are represented in pixel orientations. The MNIST is a handwritten digits dataset. The Olivettifaces dataset consists ten face images of 40 individuals with small variation in viewpoint, the addition of glass and large variation in expression. The COIL-20 is an image of 20 group objects such as animals, furniture and etc.
Table 1: Overview of datasets along with their size, dimensions, and classes
| Dataset Name | Size | Dimensions | # Classes |
| IRIS | 150 | 4 | 3 |
| Breast Cancer | 569 | 30 | 2 |
| Leukemia-ALL-AML | 72 | 7129 | 2 |
| Wine | 178 | 13 | 3 |
| MNSIT | 70K | 784 | 10 |
| Olivetti faces | 400 | 10304 | 40 |
| COIL-20 | 1440 | 1024 | 20 |
5.2 Experimental configuration
In our experimentation, The parameter k of Algorithm 1 is initialy considered as 1 ≤ k ≤ 50. The upperbound of k is equal to largest perplexity value from the literature study [9]. The perplexity is set between 5 and 50 for a fairly good visual representation of any real-world data. In our proposal, the samples of any clustering technique depends on the parameter k. Also, we observed that when there is an increment in parameter k then there is an increment or no change in FMI score of clustering until certain k value. The FMI score becomes stable afterwards. In our experimentation at most of the cases, the selected clustering technique generates representative sample with k value less than or equal to 20. In other cases, the cluster technique generates representative sample with k value greater than 20. But there is an small increment in FMI score comparatively FMI score of clustering with k value less than or equal to 20. Therefore, we have generalized the k-value as less than or equal to 20. The sensitivity of parameter k needs to be investigated more in future. The number of clusters is defined as a constant that is equal to the number of ground truth classes. The original class labels are not used anywhere in the experimental evaluation. The ground truth class labels are used only for measuring the FMI score that determines the cluster purity. The threshold parameter is set to 0.05 that provides the intuition for selecting the recommended clustering techniques from the order sequence. The threshold parameter derived from the statistical method where the maximum allowable difference between two consecutive values of either increasing or decreasing order sequence is 5%. The recommended set size and threshold parameters are inversly proposional to each other. The sensitivity of threshold parameter needs to be investigated further. From the recommended set, the best technique is chosen and the optimal sample of it being considered as the best representative sample for designing the t-SNE model. The representative sample is embedded in a 2D space using the BH-tSNE algorithm. The parameters of BH-tSNE are set up according to the paper [13] experimental setup. In addition to that, we initialized the embedding space Y by sampling the point yi from a uniform distribution with [−0.02,0.02]2 for obtaining well-separated clusters in embedding space. The datasets with more than 50 dimensions, their dimensionality is reduced to 50 by kernel PCA. The dimensionality reduction speeds up the computation of the probability distribution of the input similarity and suppresses some noise. The results of the BH-tSNE algorithm are shown in 2D scatter-plot representation. The minimum value of pij of clustered data, the early exaggeration factor α and the learning rate η values are assigned similar to the George C.linderman and Stefan Steinerberger paper.
The data points other than the representative samples are interpolated into the BH-tSNE of a representative sample using LIDWI. The parameter rx is obtained by either proposed heuristic or intuition of LION-tSNE. In the proposed heuristic dilation factor is bounded between 0 and 1. The parameter ry, ryclose and power are measured similar to LION-tSNE algorithm.
Table 2: Parameters setting for the experimental setup
| Parameter | Value |
| k of k-NN sampling | 1 |
| Threshold | 0.05 |
| Perplexity | 5 – 50 |
|
Early exaggeration coefficient Adaptive learning rate |
1 |
| Dilation Factor | 0 |
|
rx at dist perc ry at dist perc ryclose at dist perc Fixed k-value for k-NN accuracy |
3 – 10 |
Table 2 provides the parameter settings of our experimental evaluation. The parameter perplexity represents the effective number of neighbors for each data point. For instance, the small value of perplexity creates subgroups within the same cluster of t-SNE results. In contrast to small, the large value of it does not maintain the clear separation between two clusters of t-SNE results. Both cases suffer from either under-fitting or over-fitting problem that causes a lack of visual clarity. The empirical studies state that the perplexity value between 5 to 70 gives a good visual representation of t-SNE results. The parameter dist per represents the overall percentile of the representative sample that needs to be considered as inliers. For example, if we take dist per as 95th percentile, that means out of 100 points, 95 points are considered as inliers and the remaining 5 points are outliers. The parameter k plays an important role in the computation of k-NN accuracy of the data. The effect of parameter k is shown in the Figure 3. It is clear that when there is an increment in k value, accuracy decreases monotonically. The result evaluation of representative k-NN sampling-based clustering is discussed in the next section.
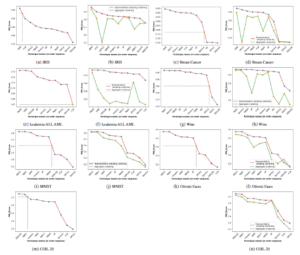
Figure 4: The column one and column three (i.e., sub-figure ((a), (c), (e), (g), (i), (k) and (m)) shows the threshold derivation (i.e., the integration of doted horizontal and vertical lines) of all seven datasets from the order sequence curve. The column two and column four (i.e., sub-figure (b), (d), (f), (h), (j), (l) and (n)) shows the relationship curve of all datasets from the order sequence, which provides the relationship between the FMI scores of proposed representative k-NN sampling-based clustering and
5.3 Result analysis
In our experimentation, we have considered most frequently and popularly used clustering techniques. The Figure 4 shows the curves of all seven datasets of subsection 5.1. The Figure 4 gives the threshold derivation for the recommended order sequence and relationship between the FMI scores of proposed representative k-NN sampling-based clustering and aggregate clustering. In the Figure 4, the x-axis represent the clustering technique names in the order sequence of FMI scores (i.e., FMI(t1) > FMI(t2) > …. > FMI(tl) where ti represents ith best clustering technique) and y-axis represents the corresponding FMI Scores. Column one and column three of Figure 4 (i.e., sub-figure (a), (c), (e), (g),(i), (k) and (m)) shows the threshold derivation for the recommendation of clustering techniques for all the given seven datasets. The column two and column four of Figure 4 (i.e., sub-figure (b), (d), (f), (h), (j), (l) and (n))
|
Table 3: The applicable clustering and their recommended order sequence for the given seven datasets.
|
shows the relationship curve that provides the relationship between the FMI Score of representative k-NN sampling-based clustering and aggregate clustering for all the given seven datasets. The Figure 4 clearly shows that the proposed representative k-NN samplingbased clustering results are superior to the aggregate clustering results.
The sets of applicable and recommended order sequence of clustering techniques for the given seven datasets is listed in the Table 3. The CP and HCI of representative k-NN sampling-based clustering and aggregate clustering of all clustering techniques are listed in the Table 4 and 5. The Table 4 and 5 clearly states that the CP of the proposed representative k-NN sampling-based clustering is smaller than the aggregate clustering for some techniques. In contrary techniques, the compactness is in reverse order, but the compactness difference is very minute in both situations. The CHI is also deriving the relationship between both clustering as same as CP where the CP is small, and then there is a large CHI score.
The contingency matrices of representative k-NN samplingbased clustering and aggregate clustering of IRIS, Wine, Breastcancer and Leukemia datasets are shown in the Table 6, 7, 8 and 9. If the number of classes of the given dataset is larger, then it is difficult to analyze the data with the contingency matrix. Therefore, the contingency matrix of MNIST, Olivetti-Faces and COIL-20 datasets are not addressed. The Table 6, 7, 8 and 9 clearly states that the proposed representative k-NN sampling-based clustering classifies the labels much similar to the ground truth class labels compared to aggregate clustering. The Tabel 9 represents the contingency matrix of the Leukemia dataset, which indicates that none of the selected technique gives the best clustering results. Still, representative k-NN sampling-based clustering produces better results than the aggregate clustering.
For Quantitative evaluation, the baseline k-NN accuracy of proposed representative k-NN sampling-based clustering is compared with baseline k-NN accuracy of aggregate clustering and baseline k-NN accuracy of ground truth class labels. The baseline k-NN accuracy and trustworthiness of representative k-NN sampling-based clustering, aggregate clustering and ground truth class labels of seven datasets are listed in Table 10. The Table 10 clearly indicates that the proposed method produces more robust results than others.
Table 4: Compactness and CHI score of representative k-NN sampling-based clustering, overall sampling clustering (i.e., assigning the cluster labels based on the KNN of remaining samples in the context of representative samples) and the aggregate clustering of IRIS, Breast-Cancer, Leukemia and Wine datasets for the selected clustering techniques.
| Method | Samp. size | Compactness | CHI Score | ||||
| Samp. cltr | Overall cltr | Aggr. cltr |
Samp. cltr |
Overall cltr | Aggr. cltr | ||
| k-Means | 73 | 0.4945 | 0.5601 | 0.5557 | 221.54 | 347.56 | 358.56 |
| Agglomerative | 63 | 0.5143 | 0.5579 | 0.5606 | 178.78 | 353.21 | 348.03 |
| Birch | 73 | 0.5291 | 0.5646 | 0.8507 | 183.85 | 335.22 | 193.97 |
| Spectral | 60 | 0.5234 | 0.5606 | 0.5709 | 167.457 | 348.03 | 322.48 |
| EMGMM | 63 | 0.5316 | 0.5754 | 0.5789 | 161.44 | 317.59 | 307.76 |
| FCM | 72 | 0.5107 | 0.5633 | 0.5576 | 197.71 | 340.79 | 355.71 |
| DBSCAN | 73 | 0.6379 | 0.7047 | 0.6723 | 104.84 | 175.96 | 124.78 |
| AP | 73 | 0.52 | 0.557 | 0.5588 | 196.99 | 354.54 | 353.22 |
| MBKM | 74 | 0.4834 | 0.5613 | 0.5562 | 234.32 | 343.95 | 356.28 |
| OPTICS | 73 | 0.5894 | 0.6071 | 1.0919 | 120.53 | 259.45 | 15.99 |
| MS | 70 | 0.527 | 0.557 | 0.6002 | 188.2 | 354.45 | 289.52 |
| Breast-Cancer | |||||||
| k-Means | 291 | 1.5608 | 1.6064 | 1.6013 | 203.91 | 355.42 | 364.09 |
| Agglomerative | 290 | 1.5735 | 1.6073 | 1.6322 | 199.73 | 356.80 | 319.01 |
| Birch | 291 | 1.5698 | 1.6083 | 1.9509 | 193.53 | 352.67 | 53.96 |
| Spectral | 289 | 1.5694 | 1.6177 | 1.6449 | 197.34 | 351.45 | 328.74 |
| EMGMM | 276 | 1.5744 | 1.6096 | 1.6168 | 178.38 | 350.08 | 337.00 |
| FCM | 276 | 1.5645 | 1.6048 | 1.6001 | 186.30 | 358.22 | 363.04 |
| DBSCAN | 291 | 1.9493 | 2.0004 | 1.9735 | 21.27 | 19.66 | 38.11 |
| AP | 213 | 1.5146 | 1.6088 | 1.6088 | 156.95 | 356.82 | 358.62 |
| MBKM | 268 | 0.8824 | 1.6047 | 1.6007 | 180.883 | 359.63 | 363.83 |
| OPTICS | 268 | 1.9635 | 2.0179 | 2.0069 | 21.72 | 24.65 | 38.41 |
| MS | 213 | 1.9138 | 2.0054 | 1.9412 | 19.79 | 17.65 | 68.97 |
| Leukemia-ALL-AML | |||||||
| k-Means | 22 | 3.4413 | 3.9367 | 3.9279 | 1.567 | 1.173 | 1.685 |
| Agglomerative | 19 | 3.3494 | 3.9315 | 3.9222 | 1.874 | 1.269 | 1.756 |
| Birch | 19 | 3.3494 | 3.9315 | 3.919 | 1.874 | 1.269 | 1.921 |
| Spectral | 19 | 3.3076 | 3.915 | 3.92 | 1.707 | 1.279 | 1.845 |
| EMGMM | 20 | 3.4012 | 3.9415 | 3.9237 | 1.337 | 0.953 | 1.591 |
| FCM | 20 | 3.3743 | 3.939 | 3.9271 | 1.622 | 1.117 | 1.619 |
| DBSCAN | 17 | 3.2654 | 3.9436 | 3.9225 | 1.351 | 0.959 | 1.326 |
| AP | 14 | 2.8993 | 3.895 | 3.9355 | 1.548 | 0.882 | 1.292 |
| MBKM | 22 | 3.3088 | 3.8957 | 3.9366 | 1.102 | 0.859 | 1.122 |
| OPTICS | 19 | 3.315 | 3.922 | 3.9071 | 0.859 | 0.717 | 1.317 |
| MS | … | …. | …. | …. | …. | …. | …. |
| Wine | |||||||
| k-Means | 82 | 1.3275 | 1.4298 | 1.4289 | 47.255 | 82.828 | 83.373 |
| Agglomerative | 87 | 1.3207 | 1.4356 | 1.4328 | 45.964 | 80.465 | 81.327 |
| Birch | 82 | 1.555 | 1.6567 | 1.629 | 19.859 | 34.395 | 42.564 |
| Spectral | 87 | 1.3532 | 1.4348 | 1.4298 | 42.372 | 81.014 | 82.828 |
| EMGMM | 71 | 1.3481 | 1.4331 | 1.4348 | 39.846 | 81.796 | 81.698 |
| FCM | 85 | 1.3473 | 1.4317 | 1.4305 | 45.029 | 82.346 | 83.135 |
| DBSCAN | 64 | 1.6181 | 1.6656 | 1.9606 | 15.719 | 37.502 | 3.102 |
| AP | 82 | 1.5534 | 1.3368 | 1.4458 | 45.362 | 80.714 | 80.828 |
| MBKM | 87 | 1.3562 | 1.4848 | 1.4278 | 42.472 | 81.784 | 81.523 |
| OPTICS | 85 | 1.4484 | 1.4531 | 1.4248 | 38.866 | 82.796 | 81.698 |
| MS | 71 | 1.3489 | 1.4371 | 1.4748 | 49.846 | 81.797 | 80.598 |
Table 5: Compactness and CHI score of representative k-NN sampling-based clustering, overall sampling clustering (i.e., assigning the cluster labels based on the KNN of remaining samples in the context of representative samples) and the aggregate clustering of MNIST Handwritten digits, Olivetti-Faces and COIL-20 datasets for the selected clustering techniques.
MNIST
| Method | Samp. size | Compactness | CHI Score | ||||
| Samp. cltr | Overall cltr | Aggr. cltr |
Samp. cltr |
Overall cltr | Aggr. cltr | ||
| k-Means | 4481 | 3816.4 | 3897.8 | 3877.8 | 233.97 | 477.45 | 492.48 |
| Agglomerative | 4900 | 3936.2 | 3974.2 | 3974.2 | 206.16 | 409.22 | 395.43 |
| Birch | 4900 | 3936.2 | 3974.2 | 3974.2 | 206.16 | 409.22 | 395.43 |
| Spectral | 4735 | 4662.2 | 4694.5 | 4695.0 | 1.048 | 1.0308 | 1.0459 |
| EMGMM | 4206 | 3862.2 | 3930.6 | 3977 | 200.8 | 443.59 | 400.32 |
| FCM | 4835 | 3962.2 | 4374.5 | 4555. | 1.248 | 1.308 | 1.59 |
| DBSCAN | 4735 | 4662.2 | 4694.5 | 4695. | 1.048 | 1.0308 | 1.0459 |
| AP | 4496 | 3832.5 | 3978.6 | 3897.3 | 213.97 | 457.78 | 472.34 |
| MBKM | 4783 | 4216.4 | 4597.1 | 4577.3 | 224.17 | 467.65 | 452.18 |
| OPTICS | 4625 | 3962.5 | 4094.3 | 4195.7 | 1.258 | 1.13 | 1.045 |
| MS | … | …. | …. | …. | …. | …. | …. |
| Olivetti-Faces | |||||||
| k-Means | 200 | 1568 | 1784.5 | 1565.8 | 10.384 | 14.009 | 21.210 |
| Agglomerative | 178 | 1495.9 | 1798.8 | 1546.8 | 10.718 | 13.634 | 22.008 |
| Birch | 178 | 1495.9 | 1798.8 | 1546.8 | 10.718 | 13.634 | 22.008 |
| Spectral | 188 | 2235 | 2545.9 | 2608.4 | 0.6858 | 0.6839 | 0.9924 |
| EMGMM | 165 | 1528.9 | 1812.8 | 1591.5 | 9.211 | 13.168 | 20.272 |
| FCM | 178 | 2479.3 | 2659.4 | 2732.6 | 4.321 | 4.874 | 4.178 |
| DBSCAN | 194 | 2679.4 | 2750.5 | 2745.1 | 4.021 | 3.804 | 5.782 |
| AP | 194 | 1628.9 | 1852.8 | 1691.5 | 10.321 | 14.168 | 20.872 |
| MBKM | 178 | 1668.6 | 1852.8 | 1671.5 | 9.711 | 12.168 | 21.275 |
| OPTICS | 165 | 1598.4 | 1932.8 | 1891.5 | 8.217 | 13.468 | 22.728 |
| MS | … | …. | …. | …. | …. | …. | …. |
| COIL-20 | |||||||
| k-Means | 679 | 10.333 | 10.362 | 10.180 | 87.911 | 183.571 | 188.454 |
| Agglomerative | 701 | 10.239 | 10.237 | 10.294 | 90.220 | 184.703 | 181.461 |
| Birch | 701 | 10.577 | 10.562 | 11.1165 | 87.377 | 180.283 | 162.538 |
| Spectral | 676 | 14.852 | 14.944 | 17.447 | 28.585 | 57.964 | 27.4288 |
| EMGMM | 679 | 10.273 | 10.243 | 10.356 | 87.832 | 186.73 | 187.686 |
| FCM | 689 | 12.872 | 13.645 | 15.745 | 34.784 | 56.768 | 34.58 |
| DBSCAN | 679 | 18.843 | 18.962 | 19.769 | 19.532 | 38.902 | 38.548 |
| AP | 701 | 10.573 | 10.253 | 10.856 | 83.83 | 188.63 | 186.656 |
| MBKM | 679 | 10.253 | 10.143 | 10.366 | 80.832 | 188.73 | 187.656 |
| OPTICS | 679 | 10.243 | 10.233 | 10.326 | 87.432 | 185.738 | 184.286 |
| MS | … | … | … | … | … | … | … |
Table 6: Contingency matrix of k-NN sampling-based and original clustering on IRIS dataset with best k-NN sampling-based clustering technique (i.e., EMGMM).
Class labels generated by
| k-NN samplingbased clustering | Aggregate clustering |
| C1 C2 C3 | C1 C2 C3 |
| 50 0 0 | 50 0 0 |
| 0 49 1 | 0 45 5 |
| 0 2 48 | 0 0 50 |
| Original |
Table 7: Contingency matrix of k-NN sampling-based and original clustering on Wine dataset with best k-NN sampling-based clustering technique (i.e., Agglomerative).
Class labels generated by
| k-NN samplingbased clustering | Aggregate clustering |
| C1 C2 C3 | C1 C2 C3 |
| 0 0 59 | 2 0 57 |
| 69 2 0 | 69 2 0 |
| 1 47 0 | 0 48 0 |
| Original |
The sampling k-NN accuracies such as train, test and overall of t-SNE embedding of a representative sample and interpolation of other samples are listed in Table 11 and the overall trustworthiness is also covered. The best representative sample of representative k-NN sampling-based clustering and random sampling of IRIS dataset is shown in Figure 5. The 2D scatter-plots shown in Figure 5 are the first two coordinates of IRIS dataset. The Figure 5(a) shows the best representative sample of k-NN sampling and the Figure 5(c) shows the representative sample of k-NN sampling along with samples of (t,m,s)-Nets of remaining data points. The Figure 5(b) and 5(d) shows the random sampling of size equal to the sample size of Figure 5(a) and 5(c). The samples of Figure 5(a) are constant and consistent comparatively other sampling.
Table 8: Contingency matrix of k-NN sampling-based and original clustering on Breast-Cancer dataset with best k-NN sampling-based clustering technique (i.e., EMGMM).
Class labels generated by
| k-NN samplingbased clustering | Aggregate clustering |
| C1 C2 | C1 C2 |
| 21 191 | 16 196 |
| 352 5 | 340 17 |
| Original |
Table 9: Contingency matrix of k-NN sampling-based and original clustering on Leukemia dataset with best k-NN sampling-based clustering technique (i.e., MBKM). Class labels generated by
| k-NN samplingbased clustering | Aggregate clustering |
| C1 C2 | C1 C2 |
| 0 47 | 9 38 |
| 2 23 | 6 19 |
| Original |
Table 10: Baseline k-NN accuracy of k-NN sampling based clustering, original clustering and ground truth class labels
|
Dataset Name |
Trust | Baseline k-NN Accuracy | ||
|
Sampling Clustering |
Aggregate Clustering | Ground-truth class labels | ||
| IRIS | 0.9861 | 0.9735 | 0.9666 | 0.96 |
| Breast-Cancer | 0.958 | 0.9876 | 0.9862 | 0.9577 |
| Leukemia | 0.6799 | 0.9875 | 0.6643 | 0.7485 |
| Wine | 0.9552 | 0.9774 | 0.9828 | 0.9717 |
| MNIST Digits | 0.9895 | 0.9259 | 0.9483 | 0.9512 |
| Olivetti face | 0.9494 | 0.6775 | 0.8287 | 0.88 |
| COIL-20 | 0.9972 | 0.9554 | 0.9709 | 0.9743 |
The Figure 6 shows the 2D scatter-plot visualization of IRIS data with outliers projection. In Figure 6, we are also showing the outliers (i.e. denoted by large grey color circle) of original data after finding the outliers from the addition of remaining data points with the radius rx which is obtained by the proposed heuristic rx = (1 + d f) ∗ rx with d f = 0.2. It clearly states that sampled data obtain the outliers of original data.
The 2D scatter-plot visualization of embedding space of representative k-NN sampling-based clustering of the MNIST dataset is shown in the Figure 7. The Figure 7 shows the baseline 2D scatter-plot of t-SNE embedding of whole MNIST data of size 10K
|
Table 11: Quantitative measure using k-NN accuracy of k-NN sampling based clustering, original clustering and ground truth class labels Optimal Sample Dataset Sample Trust Sampling k-NN Accuracy
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
with representative k-NN sampling-based clustering labels as the colors of scatter point groups.
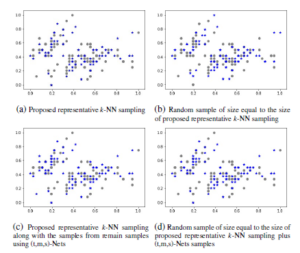
Figure 5: Training sample selection (i.e., represented by blue star scatter-plot point) from IRIS dataset with four different strategies.
The Figure 8 shows the four different 2D scatter-plot representation of MNIST dataset. In Figure 8(a), we are representing the 2D scatter-plot of sampled t-SNE along with the interpolation of inliers and placement of outliers. The sampled t-SNE is designed based on the best representative sample of the representative k-NN sampling-based clustering concerning the best clustering technique. The inliers of remaining samples (i.e, other than representative samples) are interpolated to t-SNE with the parameter rx that is obtained from the proposed heuristic. The outlier of remaining samples are placed into an existing t-SNE environment based on the Boystov et.al heuristic.
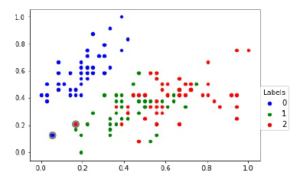
Figure 6: The 2D scatter-plot of IRIS data which denotes the outliers from the newly added sample with proposed heuristic.
In Figure 8(a), the outliers are placed separately from other embedding points. The Figure 8(b) shows the 2D scatter-plot of (t,m,s)-Nets based t-SNE along with interpolation of other samples. The (t,m,s)-Nets based t-SNE is designed with a combination of representative k-NN sampling-based clustering and (t,m,s)-Nets of remaining samples. The (t,m,s)-Nets samples are selected from the data samples, which are having NN-score as zero after selecting the samples using a mutual k-NN sampling with a static graph updation algorithm. The inliers of remaining samples (i.e, other than representative samples plus (t,m,s)-Nets samples) are interpolated to t-SNE with the parameter rx that is obtained from the Boystov et.al heuristic. The Figure 8(c) and 8(d) shows the 2D scatter-plots of a random sampled t-SNE along with the addition of new data samples. The random sample size is equal to the sample size of Figure 8(a) and 8(b). The inliers and outliers of remaining samples are placed according to the intuitions of Figure 8(a) and 8(b). The representative k-NN sampling-based results are statistically significant than the random sampling based results which is derived in our earlier paper [19]. The following section describe the conclusion and future work.
6. Conclusion and Future Work
In this paper, we have proposed a representative k-NN samplingbased clustering approach, which generates cluster results on a sampling basis. The most frequently used clustering techniques are applied to obtain the sampling-based cluster results. Initially, we determine the applicable set of techniques for the given dataset. From the applicable set, sampling-based clustering results of each technique are evaluated by an external cluster validation index called FMI-score. The applicable techniques are arranged in an orderly sequence of their FMI scores. Some threshold parameter derives the recommendation of clustering techniques for the given dataset. From the recommended set, the first technique is selected as the most desirable clustering for the given dataset.
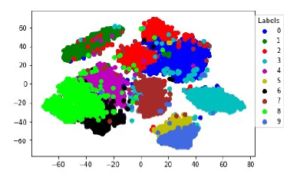
Figure 7: The baseline 2D visual representation of Representative k-NN samplingbased clustering t-SNE embedding where t-SNE applied on whole data
The samples of this technique are used for generating the lowdimensional embedding of input data. The embedding results are visualized and quantitatively evaluated in the context of samplingbased cluster results. The proposed approach is expanded to identify all the applicable set of clustering techniques for the given dataset, which can be done as future work. There is scope for implementing the proposed algorithm in a distributed environment that can be applied to Big Data.
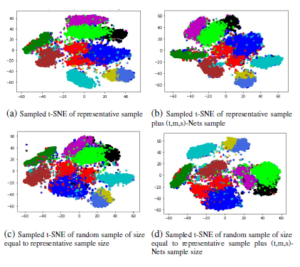
Figure 8: 2D representation of four different t-SNE embedding of MNIST dataset, (a) t-SNE embedding of representative train sample and the interpolation of inliers as well as placement of outliers with proposed heuristic, (b) t-SNE embedding of representative train sample which includes the samples from (t,m,s)-Nets and the interpolation of inliers as well as placement of outliers with A Boystov et.al heuristic, (c) t-SNE embedding of random sample of size equal to the size of sub-figure (a) and the interpolation of inliers as well as placement of outliers with proposed heuristic, (d) t-SNE embedding of random sample of size equal to the size of sub-figure (b) and the interpolation of inliers as well as placement of outliers with A Boystov et.al heuristic.
Acknowledgment
I would like to express my sincere gratitude to Visveswaraya PhD Scheme for providing financial support to pursue my PHD degree.
- Matthew Partridge, Rafael A Calvo, ”Fast dimensionality reduction and simple PCA” Intelligent data analysis, 2(3), 203–214, 1998.
- Trevor F Cox, Michael AA Cox, Multidimensional scaling, Chapman and hall/CRC, 2000.
- Jianbo Shi, J. Malik, ”Normalized cuts and image segmentation” IEEE Trans- actions on Pattern Analysis and Machine Intelligence, 22(8), 888–905, 2000, doi: 10.1109/34.868688.
- Joshua B Tenenbaum, Vin De Silva, John C Langford, ”A global geomet- ric framework for nonlinear dimensionality reduction” Science, 290(5500), 2319–2323, 2000. DOI: 10.1126/science.290.5500.2319
- Sam T Roweis, Lawrence K Saul, ”Nonlinear dimensionality reduction by locally linear embedding” Science, 290(5500), 2323–2326, 2000. DOI: 10.1126/science.290.5500.2323
- Mikhail Belkin, Partha Niyogi, ”Laplacian eigenmaps for dimensionality re- duction and data representation” Neural computation, 15(6), 1373–1396, 2003. https://doi.org/10.1162/089976603321780317
- Geoffrey E Hinton, Sam T Roweis, ”Stochastic neighbor embedding” in Ad- vances in neural information processing systems, pages 857–864, 2003.
- Laurens Van Der Maaten, Eric Postma, Jaap Van den Herik, ”Dimensionality reduction: a comparative” J Mach Learn Res, 10(66–71),13, 2009.
- Laurens Van Der Maaten, Geoffrey Hinton, ”Visualizing data using t-sne” J Mach Learn Res, 9(Nov):2579–2605, 2008.
- Wentian Li, Jane E Cerise, Yaning Yang, Henry Han, ”Application of t-sne to human genetic data” Journal of Bioinformatics and Computational Biology, 15(4):1750017, 2017. https://doi.org/10.1142/S0219720017500172
- Minh Nguyen, Sanjay Purushotham, Hien To, Cyrus Shahabi, ”m-tsne: A framework for visualizing high-dimensional multivariate time series” arXiv preprint arXiv:1708.07942, 2017.arXiv:1708.07942
- Walid M Abdelmoula, Benjamin Balluff, Sonja Englert, Jouke Dijkstra, Marcel JT Reinders, Axel Walch, Liam A McDonnell, Boudewijn PF Lelieveldt, ”Data-driven identification of prognostic tumor subpopulations using spatially mapped t-sne of mass spectrometry imaging data” in Pro- ceedings of the National Academy of Sciences, 113(43):12244–12249, 2016. https://doi.org/10.1073/pnas.1510227113
- Laurens Van Der Maaten, ”Accelerating t-sne using tree-based algorithms” J Mach Learn Res, 15:3221–3245, 2014.
- Nicola Pezzotti, Boudewijn PF Lelieveldt, Laurens Van Der Maaten, Thomas Ho¨llt, Elmar Eisemann, Anna Vilanova, ”Approximated and user steerable tsne for progressive visual analytics” IEEE transactions on visualization and com- puter graphics, 23(7):1739–1752, 2016. DOI: 10.1109/TVCG.2016.2570755
- Laurens Van Der Maaten, ”Learning a parametric embedding by preserving local structure” in Artificial Intelligence and Statistics, pages 384–391, 2009.
- Andrej Gisbrecht, Alexander Schulz, Barbara Hammer, ”Parametric nonlinear dimensionality reduction using kernel t-sne” Neurocomputing, 147:71–82, 2015. https://doi.org/10.1016/j.neucom.2013.11.045
- Andrey Boytsov, Francois Fouquet, Thomas Hartmann, Yves LeTraon, ”Visu- alizing and exploring dynamic high-dimensional datasets with lion-tsne” arXiv preprint arXiv:1708.04983, 2017. arXiv:1708.04983
- Stephen Ingram, Tamara Munzner, ”Dimensionality reduction for docu- ments with nearest neighbor queries” Neurocomputing, 150:557–569, 2015. https://doi.org/10.1016/j.neucom.2014.07.073
- Bheekya Dharamsotu, K. Swarupa Rani, Salman Abdul Moiz, C. Raghaven- dra Rao, ”k-NN sampling for visualization of dynamic data using LOIN- tSNE” in 2019 IEEE 26th International Conference on High Perfor- mance Computing, Data, and Analytics (HiPC), pages 63–72, 2019. DOI: 10.1109/HiPC.2019.00019
- L. J. Williams, H. Abdi, “Fishers least significant difference (lsd) test,” Ency- clopedia of research design, 218, 840–853, 2010.
- Tapas Kanungo, David M Mount, Nathan S Netanyahu, Christine D Piatko, Ruth Silverman, Angela Y Wu, ”An efficient k-means clustering algorithm: Analysis and implementation” IEEE Transactions on Pattern Analysis and Ma- chine Intelligence, 24(7):881–892, 2002. DOI: 10.1109/TPAMI.2002.1017616
- Wei Zhang, Deli Zhao, Xiaogang Wang, ”Agglomerative clustering via maxi- mum incremental path integral” Pattern Recognition, 46(11):3056–3065, 2013. https://doi.org/10.1016/j.patcog.2013.04.013
- Tian Zhang, Raghu Ramakrishnan, Miron Livny, ”BIRCH: an efficient data clustering method for very large databases” ACM Sigmod Record, 25(2):103– 114, 1996. https://doi.org/10.1145/235968.233324
- James C. Bezdek, Robert Ehrlich, William Full, ”FCM: The fuzzy c-means clustering algorithm” Computers and Geosciences, 10(2):191–203, 1984.
- Erich Schubert, Jo¨ rg Sander, Martin Ester, Hans Peter Kriegel, Xiaowei Xu, ”DBSCAN revisited, revisited: why and how you should (still) use DB- SCAN” ACM Transactions on Database Systems (TODS), 42(3):1–21, 2017. https://doi.org/10.1145/3068335
- John Carlo Dassun, Argie Reyes, Hideo Yokoyama, P Bibangco El Jireh, Mehreen Dolendo, ”Ordering points to identify the clustering structure algo- rithm in fingerprint-based age classification” Virtutis Incunabula, 2(1):17–27, 2015.
- Dorin Comaniciu, Peter Meer, ”Mean shift: A robust approach toward feature space analysis” IEEE Transactions on Pattern Analysis and Machine Intelli- gence, 24(5):603–619, 2002. DOI: 10.1109/34.1000236
- Ulrike Von Luxburg, ”A tutorial on spectral clustering” Statistics and Comput- ing, 17(4):395–416, 2007.
- Douglas A Reynolds, ”Gaussian mixture models” Encyclopedia of biometrics, 741, 2009.
- Brendan J. Frey, Delbert Dueck, ”Clustering by passing messages between data points” Science, 315(5814):972–976, 2007. DOI: 10.1126/science.1136800
- David Sculley, ”Web-scale k-means clustering” in Proceedings of the 19th international conference on World Wide Web, pages 1177–1178, 2010. https://doi.org/10.1145/1772690.1772862
- Edward B Fowlkes, Colin L Mallows, ”A method for comparing two hierarchi- cal clusterings” Journal of the American Statistical Association, 78(383):553– 569, 1983.
- Y. Liu, Z. Li, H. Xiong, X. Gao, J. Wu, S. Wu, ”Understanding and en- hancement of internal clustering validation measures” IEEE Transactions on Cybernetics, 43(3):982–994, 2013. DOI: 10.1109/TSMCB.2012.2220543
- Shusaku Tsumoto, ”Contingency matrix theory: Statistical dependence in a contingency table” Information Sciences, 179(11):1615–1627, 2009. https://doi.org/10.1016/j.ins.2008.11.023
- Amit Saxena, Mukesh Prasad, Akshansh Gupta, Neha Bharill, Om Prakash Patel, Aruna Tiwari, Meng Joo Er, Weiping Ding, Chin-Teng Lin, ”A review of clustering techniques and developments” Neurocomputing, 267:664–681, 2017. https://doi.org/10.1016/j.neucom.2017.06.053
- Anil K Jain, M Narasimha Murty, Patrick J Flynn, ”Data cluster- ing: a review” ACM computing surveys (CSUR), 31(3):264–323, 1999. https://doi.org/10.1145/331499.331504
- Pavel Berkhin, ”A survey of clustering data mining techniques” in Grouping multidimensional data, pages 25–71. Springer, 2006.
- Andrew Y Ng, Michael I Jordan, Yair Weiss, ”On spectral clustering: Analysis and an algorithm” in Advances in neural information processing systems, pages 849–856, 2002.
- George C Linderman, Stefan Steinerberger, ”Clustering with t-sne, prov- ably” SIAM Journal on Mathematics of Data Science, 1(2):313–332, 2019. https://doi.org/10.1137/18M1216134
- Donald Shepard, ”A two-dimensional interpolation function for irregularly- spaced data” in Proceedings of the 1968 23rd ACM national conference, pages 517–524, 1968. https://doi.org/10.1145/800186.810616
- Jarkko Venna, Samuel Kaski, ”Neighborhood preservation in nonlinear projec- tion methods: An experimental study” in International Conference on Artificial Neural Networks, pages 485–491. Springer, 2001.
- T. Kollig, A. Keller, ”Efficient multidimensional sampling” Computer Graphics Forum, 21(3), 2002.
- Dheeru Dua, Casey Graff, UCI machine learning repository, 2019.
- Bernhard Scho¨lkopf, Alexander Smola, Klaus-Robert Mu¨ller, ”Kernel principal component analysis” in International conference on artificial neural networks,
pages 583–588. Springer, 1997.
