A Modular Multipurpose, Parameter Centered Electronic Health Record Architecture
Volume 4, Issue 5, Page No 334-340, 2019
Author’s Name: Norbert Maggi1,2, Luca Douglas Magnoni2, Carmelina Ruggiero1,2, Roberta Gazzarata2, Mauro Giacomini1,2,a)
View Affiliations
1Department of Informatics, Bioengineering, Robotics and Systems Engineering (DIBRIS), University of Genova, Genova, Italy
2Healthropy S.r.l., Savona, Italy
a)Author to whom correspondence should be addressed. E-mail: mauro.giacomini@dibris.unige.it
Adv. Sci. Technol. Eng. Syst. J. 4(5), 334-340 (2019); ![]() DOI: 10.25046/aj040543
DOI: 10.25046/aj040543
Keywords: EHR, HL7, Interoperability
Export Citations
Health Information Technology is playing a key role in healthcare. Specifically, the use of electronic health records has been found to bring about most significant improvements in healthcare quality, mainly as relates to patient management, healthcare delivery and research support. Health record systems adoption has been promoted in many countries to support efficient, high quality integrated healthcare. The objective of this work is the implementation of an Electronic Health Record system based on a relational database. The system architecture is modular and based on the concentration of specific pathology related parameters in one module, therefore the system can be easily applied to different pathologies. Several examples of its application are described. It is intended to extend the system integrating genomic data.
Received: 15 June 2019, Accepted: 06 September 2019, Published Online: 22 October 2019
1. Introduction
This paper is an extension of work originally presented in IEEE 4th International Forum on Research and Technology for Society and Industry [1]
The collection of health information for patient data management including patient characteristics, treatments and outcomes has been over the years a most important aspect of health care [2, 3]. EHRs, which replace paper medical records to collect people’s health information, integrate different and heterogeneous data and are one of the most widely used Health Information Technology (HIT) tools. HIT contributes to the improvement of quality of care, to patient’s safety and to the reduction of health care costs by making available accurate documentation and rapid information retrieval and management. The adoption of EHRs has been promoted in many countries, both at government level an in the private sector, even though their adoption is still rather limited, mostly because of standardization, certification, security and privacy aspects and concerns. The adoption of standard based EHR systems has been found to be able to reduce spending for healthcare to a great extent [4]. EHRs can improve patient care and safe practice, however potential risks related to medical error, systems failure, and security and legal responsibility aspects should be considered, identifying barriers for EHRs adoption and ways to address them [5-9]. Moreover, organizational and human factors can promote or hamper the adoption of a EHR [10]. Further relevant aspects of EHRs are interfaces with other information systems, that should be based on standard formats, such as Health Level 7 Clinical Document Architecture (HL7 CDA), HL7 Version 3, openEHR and on controlled vocabularies such as Logical Observation Identifiers Names and Codes (LOINC), “International Classification of Diseases (ICD), Systematized Nomenclature of Medicine (SNOMED) and others [11]. The need for generic and interoperable EHRs has been recognized, specially as relates to interoperability, for the preservation of clinical information across heterogeneous systems [12]. Standards are needed to implement interoperability between systems.
An EHR system contains quantitative data (clinical data), qualitative data (text documents), and medication records. The related amount of data is rapidly expanding, and big data technology is going to play a key role in this respect, specially as relates to natural language processing, clinical decision support, integration of genomics and system biology with EHR data, knowledge dissemination and interaction with patients [13].
The set of requirements that must be met by EHR systems is defined by ISO 18308:2011. According to this standard the scope of the EHR is regarded as broader than documentation, prevention and treatment of illnesses. ISO 18308 relates to shared EHR information and to the EHR system aspects that may be used for the support and coordination of patient-centred continuity of care. An EHR system includes data repositories, directory services, knowledge services containing terminological system, care pathways and workflows, user applications and other modules and services. An EHR architecture can be classified according to a data approach, a concepts approach and a process/service approach. These approaches are present in existing systems and in specific domain languages and modelling languages [14]. A medical record system has been developed, adopting the three approaches above. The medical record system that has been developed is based on Health Level 7 (HL7) standard and can be easily tailored to the needs of different clinical wards. The use of this standard allows the system to support interoperability between the main components of the Hospital Information System (HIS), such as the Laboratory Information System (LIS) and the Pharmaceutical Information System (PIS), and can be easily extended for example to the radiology information system (RIS) and to the picture archiving and communication system (PACS). A Service Oriented Architecture (SOA) has been adopted, that can be defined as an open extensible, composable architecture based on autonomous, interoperable and reusable services, implemented as a Web services [15]. SOA has been employed in healthcare after adoption in other sectors, such as banking, transport and industrial automation [16]. SOA is widely used for real time implementation in large distributed systems, mostly because it supports the easy integration of new software within the existing one, reducing the impact on service users and cost [17-20]. Moreover, the SOA paradigm has been successfully implemented in the design of several distributed healthcare systems [21-23].
2. Materials and Methods
The system has been implemented taking into account the specific needs of different departments of the San Martino hospital in Genoa (Italy).
The workflow relating to a visit includes the identification of the patient through the information already present in the HIS obtained by a pseudo-anonym code according to the specifications of UE General Data Protection Regulation, (GDPR) [24]. The information on laboratory analyses is obtained by query of the hospital LIS and the information on therapies is obtained by query of the PIS of the hospital. The data are stored on different platforms which do not interact with each other, therefore its direct use is cumbersome and with possible misunderstandings. In order to address this problem, a system based on a relational database developed with Microsoft SQL Server 2017 has been set up, which has made it possible to create a consistent and logical representation of the information. It is worth noting that, in order to achieve GDPR compliance, the data in the database are stored by pseudo-anonymization techniques. A pseudo-anonymous code is used to query the database, in a secure fashion. The information relating to the identity of the patient is present only at the physician interface level, after authentication [23].
The database has been designed developing a conceptual model of the data, abstracting relevant aspects of the activities and processes relating to patient management. A conceptual model is a set of rules and conventions that allows to move from a reality of interest to a conceptual scheme of the data. The model has been divided into three levels that represent respectively the protagonists of the project (level 0), the clinical event (level 1), and the format of the parameters and results (level 2).
In level 0 the main actors and features of the project have been considered, defining the fundamental entities and the relationships among them. The Entity-Relation (E-R) diagram shows the general logistical aspects of the platform (Figure 1). Patients are followed up and subsequently entered into the database by the operators. Both patients and operators are linked to the centres involved in each clinical study. Each operator may be authorised to access more than one clinical study, and each patient can be involved in more than one study. Moreover, the clinical events relating to each patient may take place in different centres (the latter aspect being of fundamental importance, since different centres may use different units of measurement and ranges of normality). Each study may consider more than one clinical event, some of which may be common to those of other studies, while others may be specific to certain studies only.
 Figure 1. Entity relationship diagram for level 0
Figure 1. Entity relationship diagram for level 0
Level 1 of the E-R diagram (Figure 2) shows the meta-descriptive approach that has been adopted to manage clinical events and make the database structure as simple and general as possible. The main element is the definition of different Event Types. Within the conceptual definition of systems implemented with E-R diagrams [25], attributes are the elements that describe in detail the characteristics of entities and relationships. By transforming E-R diagrams with relational logic schemes, these attributes become columns of tables in databases. This approach, if applied to our case, binds the set of elements considered in a medical record (which happens in many third-party applications), blocking it in the data base (DB) construction.
When developing the system, it has been chosen to store the names of the elements that are regarded as rows of the table (called “parameters”). Another table (called “categories”) has been categorized, and related with the clinical reality through the links in the table that derive from the relation shown in in figure 2 (diamond “considered aspect divided by”). This choice allows to use the same DB and the same software for several tables and to easily characterize the individual parameters in a strong semantic way (using international codes such as – LOINC – ICD – … ). This greatly helps interoperability.
The most important aspects have been divided into different “Categories”. These are “containers” of parameters, homogeneous from the conceptual point of view, but not necessarily homogeneous from a data format point of view (for example, a clinical event of the “Therapy” type has a “Therapy information” category, including parameters such as “Adherence” and “Therapy start date”). Each clinical event has Results, which are the values of the parameters of that particular event.
Events are grouped into types. Parameters are also grouped into types, which are translated and mapped in LOINC using standardized dictionaries, in order to support data reuse.
 Figure 2. Entity relationship diagram for level 1
Figure 2. Entity relationship diagram for level 1
Level 2 shows the entities and relationships which are taken into account to manage the Parameters and Results of Clinical Events. Each Parameter has a specific format (typically real integer, string or Boolean), so the results (i.e. the records corresponding to the values of the parameters of an event) can have different formats. The E-R diagram (Figure 3) takes into account the fact that all parameters stored in the database have their own units of measurement and range of normality, but these units and ranges are different for each centre that may be involved in the network. This is of fundamental importance for the work carried out by the physician. In the transition from the conceptual scheme to the logical scheme, great attention has been paid to this aspect, in order to allow each user to work with the tools that are most familiar to him/her. Moreover, the differences between the units of measurement have been taken into account in the data extraction algorithm. All records relating to a single parameter are converted into a standard unit of measurement, defined by the users as the ones which are most widely used.
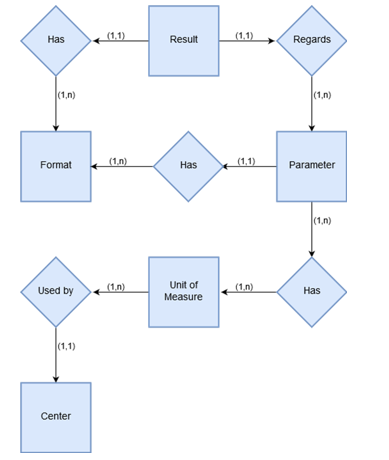 Figure 3. Entity relationship diagram for level 2
Figure 3. Entity relationship diagram for level 2
The implementation, including the creation of a web client and a web service, has been carried out in Visual Basic .NET using Microsoft Visual Studio 2015 software.
The system architecture has been designed with special reference to interoperability, according to HL7 in the HSSP (Healthcare Services Specification Project). The HSSP, started in 2005 by HL7, focuses on standard software services for healthcare, enabling the use of SOA, which has been recognised as extremely useful for interoperability, code reuse and new needs response [26]. Standards service interfaces for infrastructure reuse capability and specific health facility were considered providing a methodology for the implementation of relevant services [27].
Specifically, the HL7 Clinical Document Architecture (CDA) has been used for the encapsulation of clinical data that can be easily exchanged between applications, retaining their semantic significance [28].
Clinical parameters are associated differently with specific names and internal codes in different hospital and/or health centres. In order to allow the communication among different centres, the codes used by different hospitals/centres have been translated into LOINC language with regard to clinical trials [29], international Anatomical Therapeutic Chemical Classification System (ATC), national Marketing Authorisation (AIC) for drugs, International Classification of Diseases 9th version (ICD9) for diagnosis and ISO Country 3166-1 for the classification of the country of origin of patients.
|
Table 1. Clinical events for the Maculopathies, HIV, and Tuberculosis
|
The “San Martino” hospital in Genoa is equipped with a network infrastructure made of two Local Area Networks (LANs) that are not connected to each other (the hospital LAN and the university LAN). Specifically, the parts of the HIS are placed in the hospital LAN, which is not accessible from external networks for security reasons, because of the hospital firewall. The access to data and to the required information is granted by a virtual private network (VPN), capable to securely connect the two LANs, allowing access for clients belonging to the university network.
The original data are sent through the VPN and, by tunnelling protocols and advanced encryption, are secured by a protective barrier before they are shipped over a potentially dangerous infrastructure such as the Internet. Tunnelling is a multi-protocol (IPsec) data encapsulation process within another IP packet that is shipped over the network. In order to transmit the data to authorized end recipients only, the data are packaged twice by a specific authentication system in which access levels depend on user role.
The strong modularity of the system allows to use the general structure described above for different diseases. The system has so far been used for the implementation of the EHR for the Ligurian HIV network [30-32], for ophthalmology [33, 34] and for infectious diseases [35]. For each of these cases, different clinical events, parameter categories and parameters have been implemented in order to provide a tool which is very suited to the specific needs of physicians and caregivers. Table 1 and table 2 show, respectively, the clinical events and the categories of parameters for the three scenarios.
3. Results
The system that has being developed is hosted in Genoa university network. The user interface of the system is divided into several parts, each concerning a specific section of the medical visit. The EHR system is managed through a web interface that, for security reason, can only be accessed within the Genoese hospital and university LANs. The system interaction with the physician or caregiver takes place as follows. When the user logs in he/she is redirected to the page relating to the specific patient selection and the examination starts. After obtaining access permission, the caregiver can either manually enter the data of a new patient or see the data for a patient who is already present in the data base.
For a new entry, the related information is retrieved from the hospital databases and, after translation into LOINC, is stored in the EHR system database. The clinical parameters are retrieved from the LIS and the information on the laboratory tests is stored by an array of CDA which is sent to the web service which stores it in the system database.
A section of the clinical diary allows physicians to inspect the exams that are available for each patient. The physician can see all specific visits, their dates and parameters. The information for each clinical event is retrieved and updated by algorithms that taking into account the ethical committee permissions.
The creation of the web pages for user interface is also guided by the structure of the DB, i.e. a page is formed for each event, within which all parameters related to that event are presented. The type of value for each parameter (integer, real, list or free text, date) automatically inserts the selection of the most suitable input tool for that type (textbox, dropdown list, calendar, …). The measurement units for the numerical parameters and the normal ranges also (possibly linked to the patient’s personal data, age, sex, height, weight) are also automatically inserted.
|
Table 2. Categories for Maculopathies, HIV, and Tuberculosis
|
|
Table 3. Use of EHRs for pathologies
|
4. Discussion and Conclusions
An EHR system has been set up in order to meet the needs of medical and clinical personnel. The main aspects that have been considered relate to the management of visits and to the setting up of a clinical diary according to the standards currently in use.
The objectives of the study have been met as shown in table 3, in which the system’s application to maculopathies, HIV, and tuberculosis are described. Specifically, for the three pathologies the table shows the initial year, the numbers of patients which have been recorded, the numbers of related parameters, the number of events and the number of values that have been stored. The last column, named “session”, shows the number of days in which an operator queries an EHR.
The system aims to simplify physician’s, nurses’ and caregiver’s work for administration and management of treatment and follow-up. Moreover, it aims to support multicentric tracking for patients who are treated in several hospital facilities and the conduction of clinical trials. The system was set up focusing on parameters and parameter types, grouping them in one specific module. This facilitates the use of the system for different pathologies, because the pathology related data are in one module per pathology. The use of one single section for the management of different pathologies allows for easy use by different departments of one hospital and by different hospitals. This design choice also facilitates data reuse.
The main problems and limitation that have been encountered relate to data availability, because of data anonymity requirements for security and privacy. In the first place, this problem has been circumvented requiring the hospital to make the data pseudo-anonym, while keeping the link between the pseudo anonym code and the patient identity separate from the data and managed by the hospital system only, on physically separated platform. Moreover, the data which are copied onto the EHR system platform are encrypted by up to date techniques which protect them from attacks. Therefore, even if the data were red this would not provide any useful information
It is intended to evaluate the system performance by questionnaires addressing usability by physician, nurses and care giver.
Further developments mostly relate to the integration of genomic data in the system [36, 37]. This aspect is becoming increasingly important and the parameter focused architecture of the system is suitable in this respect. The integration of genotype and phenotype information in the EHR and the insertion of genomic information in clinical workflows would facilitate the planning, administration and evaluation of genetically enabled care.
- N. Maggi, C. Ruggiero, M. Giacomini, “Towards an Electronic Medical Record for Precision Oncology: Information and Knowledge Management,” in 2018 IEEE 4th International Forum on Research and Technology for Society and Industry (RTSI), 2018. https://doi.org/10.1109/rtsi.2018.8548422
- B. Blobel, Analysis, design and implementation of secure and interoperable distributed health information systems. Amsterdam ; Oxford: IOS Press, 2002.
- P. Coorevits, M. Sundgren, G. O. Klein, A. Bahr, B. Claerhout, C. Daniel, M. Dugas, D. Dupont, A. Schmidt, P. Singleton et al., “Electronic health records: new opportunities for clinical research,” Journal of Internal Medicine, 274(6), 547-560, 2013. https://doi.org/10.1111/joim.12119
- R. Hillestad, J. Bigelow, A. Bower, F. Girosi, R. Meili, R. Scoville, R. Taylor, “Can Electronic Medical Record Systems Transform Health Care? Potential Health Benefits, Savings, And Costs,” Health Affairs, 24(5), 1103-1117, 2005. https://doi.org/10.1377/hlthaff.24.5.1103
- V. Palabindala, A. Pamarthy, N. R. Jonnalagadda, “Adoption of electronic health records and barriers,” Journal of Community Hospital Internal Medicine Perspectives, 6(5), 2016. https://doi.org/10.3402/jchimp.v6.32643
- J. M. Gesulga, A. Berjame, K. S. Moquiala, A. Galido, “Barriers to Electronic Health Record System Implementation and Information Systems Resources: A Structured Review,” Procedia Computer Science, 124(544-551, 2017. https://doi.org/10.1016/j.procs.2017.12.188
- S. Ajami, T. BagheriTadi, “Barriers for Adopting Electronic Health Records (EHRs) by Physicians,” Acta Informatica Medica, 21(2), 2013. https://doi.org/10.5455/aim.2013.21.129-134
- A. Boonstra, M. Broekhuis, “Barriers to the acceptance of electronic medical records by physicians from systematic review to taxonomy and interventions,” BMC Health Services Research, 10(1), 2010. https://doi.org/10.1186/1472-6963-10-231
- C. M. DesRoches, D. Charles, M. F. Furukawa, M. S. Joshi, P. Kralovec, F. Mostashari, C. Worzala, A. K. Jha, “Adoption Of Electronic Health Records Grows Rapidly, But Fewer Than Half Of US Hospitals Had At Least A Basic System In 2012,” Health Affairs, 32(8), 1478-1485, 2013. https://doi.org/10.1377/hlthaff.2013.0308
- P. C. Tang, J. S. Ash, D. W. Bates, J. M. Overhage, D. Z. Sands, “Personal Health Records: Definitions, Benefits, and Strategies for Overcoming Barriers to Adoption,” Journal of the American Medical Informatics Association, 13(2), 121-126, 2006. https://doi.org/10.1197/jamia.M2025
- C. J. McDonald, “The barriers to electronic medical record systems and how to overcome them,” J Am Med Inform Assoc, 4(3), 213-21, May-Jun 1997.
- D. Kalra, “Electronic Health Record Standards,” Yearbook of Medical Informatics, 15(01), 136-144, 2018. https://doi.org/10.1055/s-0038-1638463
- T. B. Murdoch, A. S. Detsky, “The Inevitable Application of Big Data to Health Care,” Jama, 309(13), 2013. https://doi.org/10.1001/jama.2013.393
- B. Blobel, “Interoperable EHR Systems–Challenges, Standards and Solutions,” European Journal for Biomedical Informatics, 14(2), 10-19, 2018.
- T. Erl, Service-Oriented Architecture: Concepts, Technology, and Design. Pearson Education, Limited, 2016.
- S. R. Loya, K. Kawamoto, C. Chatwin, V. Huser, “Service Oriented Architecture for Clinical Decision Support: A Systematic Review and Future Directions,” Journal of Medical Systems, 38(12), 2014. https://doi.org/10.1007/s10916-014-0140-z
- R. Gazzarata, F. Vergari, T. S. Cinotti, M. Giacomini, “A standardized SOA for clinical data interchange in a cardiac telemonitoring environment,” IEEE J Biomed Health Inform, 18(6), 1764-74, Nov 2014. https://doi.org/10.1109/JBHI.2014.2334372
- E. Vasilescu, S. K. Mun, “Service Oriented Architecture (SOA) implications for large scale distributed health care enterprises,” in 1st Transdisciplinary Conference on Distributed Diagnosis and Home Healthcare, Conference Proceedings, 2006. https://doi.org/10.1109/Ddhh.2006.1624805
- R. Gazzarata, B. Giannini, M. Giacomini, “A SOA-Based Platform to Support Clinical Data Sharing,” Journal of Healthcare Engineering, 2017(Article ID 2190679), 1-24, 2017. https://doi.org/10.1155/2017/2190679
- R. Gazzarata, M. Giacomini, “A Standardized SOA for Clinical Data Sharing to Support Acute Care, Telemedicine and Clinical Trials,” European Journal for Biomedical Informatics, 12(01), 2016. https://doi.org/10.24105/ejbi.2016.12.1.9
- H. Kondylakis, B. Claerhout, M. Keyur, L. Koumakis, J. van Leeuwen, K. Marias, D. Perez-Rey, K. De Schepper, M. Tsiknakis, A. Bucur, “The INTEGRATE project: Delivering solutions for efficient multi-centric clinical research and trials,” J Biomed Inform, 62(Aug 2016), 32-47, 2016. https://doi.org/10.1016/j.jbi.2016.05.006
- R. Alonso-Calvo, S. Paraiso-Medina, D. Perez-Rey, E. Alonso-Oset, R. van Stiphout, S. Yu, M. Taylor, F. Buffa, C. Fernandez-Lozano, A. Pazos et al., “A semantic interoperability approach to support integration of gene expression and clinical data in breast cancer,” Comput Biol Med, 87(Aug 2017), 179-186, 2017. https://doi.org/10.1016/j.compbiomed.2017.06.005
- G. Gazzarata, R. Gazzarata, M. Giacomini, “A standardized SOA based solution to guarantee the secure access to EHR,” Conference on Enterprise Information Systems/International Conference on Project Management/Conference on Health and Social Care Information Systems and Technologies, Centeris/Projman / Hcist 2015, 64(1124-1129, 2015. https://doi.org/10.1016/j.procs.2015.08.582
- Regulation (EU) 2016/679 of the European Parliament and of the Council of 27 April 2016 on the protection of natural persons with regard to the processing of personal data and on the free movement of such data, and repealing Directive 95/46/EC (General Data Protection Regulation), 2016.
- A. Silberschatz, H. F. Korth, S. Sudarshan, Database system concepts, 6th ed. ed. New York: McGraw-Hill Higher Education ; London : McGraw-Hill [distributor], 2011.
- M. W. Bridges, “SOA in healthcare, Sharing system resources while enhancing interoperability within and between healthcare organizations with service-oriented architecture,” Health Management Technology, 28(6), 6, 8, 10-6, 8, 10, 2007.
- K. Kawamoto, A. Honey, K. Rubin, “The HL7-OMG Healthcare Services Specification Project: motivation, methodology, and deliverables for enabling a semantically interoperable service-oriented architecture for healthcare,” J Am Med Inform Assoc, 16(6), 874-81, Nov-Dec 2009. https://doi.org/10.1197/jamia.M3123
- R. H. Dolin, L. Alschuler, S. Boyer, C. Beebe, F. M. Behlen, P. V. Biron, A. Shabo Shvo, “HL7 Clinical Document Architecture, Release 2,” J Am Med Inform Assoc, 13(1), 30-9, Jan-Feb 2006. https://doi.org/10.1197/jamia.M1888
- Logical Observation Identifiers Names and Codes (LOINC). Available: http://loinc.org
- P. Fraccaro, V. Pupella, R. Gazzarata, C. Dentone, G. Cenderello, P. De Leo, F. Bozzano, G. Casalino Finocchio, A. De Maria, D. Fenoglio et al., “The Ligurian Human Immunodeficiency Virus Clinical Network: A Web Tool to Manage Patients With Human Immunodeficiency Virus in Primary Care and Multicenter Clinical Trials,” Medicine 2.0, 2(2), 2013. https://doi.org/10.2196/med20.2712
- B. Giannini, N. Riccardi, G. Cenderello, A. Di Biagio, C. Dentone, M. Giacomini, “From Liguria HIV Web to Liguria Infectious Diseases Network: How a Digital Platform Improved Doctors’ Work and Patients’ Care,” AIDS Research and Human Retroviruses, 34(3), 239-240, 2018. https://doi.org/10.1089/aid.2017.0064
- M. Bonetto, S. Mora, N. Maggi, C. Ruggiero, M. Giacomini, “Standards for the Reuse of Clinical Data in Research: the Connection of the Liguria HIV Network and the CISAI Cohort,” in 2019 Global Medical Engineering Physics Exchanges/ Pan American Health Care Exchanges (GMEPE/PAHCE), 2019. https://doi.org/10.1109/gmepe-pahce.2019.8717316
- P. Fraccaro, M. Nicolo, M. Bonetto, M. Giacomini, P. Weller, C. E. Traverso, M. Prosperi, D. Osullivan, “Combining macula clinical signs and patient characteristics for age-related macular degeneration diagnosis: a machine learning approach,” BMC Ophthalmology, 15(1), 2015. https://doi.org/10.1186/1471-2415-15-10
- M. Bonetto, M. Nicolo, R. Gazzarata, P. Fraccaro, R. Rosa, D. Musetti, M. Musolino, C. E. Traverso, M. Giacomini, “I-Maculaweb: A Tool to Support Data Reuse in Ophthalmology,” IEEE Journal of Translational Engineering in Health and Medicine, 4(1-10, 2016. https://doi.org/10.1109/jtehm.2015.2513043
- N. Riccardi, B. Giannini, M. L. Borghesi, L. Taramasso, E. Cattaneo, G. Cenderello, F. Toscanini, M. Giacomini, E. Pontali, G. Cassola et al., “Time to change the single-centre approach to management of patients with tuberculosis: a novel network platform with automatic data import and data sharing,” ERJ Open Research, 4(1), 2018. https://doi.org/10.1183/23120541.00108-2017
- N. Maggi, R. Gazzarata, C. Ruggiero, C. Lombardo, M. Giacomini, “Cancer precision medicine today: Towards omic information in healthcare systems,” Tumori Journal, 105(1), 38-46, 2018. https://doi.org/10.1177/0300891618792473
- N. Maggi, L. M. Douglas, C. Ruggiero, R. Gazzarata, M. Giacomini, “Information Technology System Including Patient Generated Health Data for Cancer Clinical Care and Research,” Studies in health technology and informatics, 261(289-293, 2019.
