Melanoma detection using color and texture features in computer vision systems
Volume 4, Issue 5, Page No 16-22, 2019
Author’s Name: Antonio Fuduli1, Pierangelo Veltri2, Eugenio Vocaturo3, Ester Zumpano3,a)
View Affiliations
1Department of Mathematics and Computer Science, University of Calabria, 87036, Italy
2DSCM – Bioinformatics Laboratory Surgical and Medical Science Department, University Magna Graecia, 88100, Italy
3DIMES – Department of Computer Science, Modelling, Electronic and Systems Engineering, University of Calabria, 87036, Italy
a)Author to whom correspondence should be addressed. E-mail: zumpano@dimes.unical.it
Adv. Sci. Technol. Eng. Syst. J. 4(5), 16-22 (2019); ![]() DOI: 10.25046/aj040502
DOI: 10.25046/aj040502
Keywords: Melanoma detection, Feature selection, Classification
Export Citations
All forms of skin cancer are becoming widespread. These forms of cancer, and melanoma in particular, are insidious and aggressive and if not treated promptly can be lethal to humans. Effective treatment of skin lesions depends strongly on the timeliness of the diagnosis: for this reason, artificial vision systems are required to play a crucial role in supporting the diagnosis of skin lesions. This work offers insights into the state of the art in the field of melanoma image classification. We include a numerical section where a preliminary analysis of some classification techniques is performed, using color and texture features on a data set constituted by plain photographies, to which no pre-processing technique has been applied. This is motivated by the necessity to open new horizons in creating self-diagnosis systems for accessible skin lesions, due also to a huge innovation of cameras, smartphones technology and wearable devices.
Received: 14 June 2019, Accepted: 22 August 2019, Published Online: 06 September 2019
1. Introduction
The World Health Organization recognizes skin cancer as one of the most lethal cancers that is spreading widely in many parts of the world in recent decades. Globally, each year between 2 and 3 million of non-melanoma skin cancers and 132,000 of melanoma skin cancers occur [1]. Cutaneous melanoma is still incurable. A successful treatment of skin lesion is heavily dependent on the timeliness of the diagnosis: it is crucial to detect and remove the skin lesion in the early stage. One of the major allert of this tumor is that, in the early stages, melanoma remains hidden, showing itself to be similar to other benign lesions. This aspect, combined with the aggressiveness and rapidity with which it expands, creates difficulties even for expert dermatologists; in addition, in its early phase, it is often underestimated [2]. Dermatologists, in clinical evaluation, in order to identify the tumor lesions of the skin, adhere to some reference protocols. Generally, they refer to the ABCDE protocol [3], the 7-point checklist [4] and the Menzies method [5]. If the investigations conducted leave doubts about the nature of the lesion, the specialist prescribes an analysis of the affected tissue by biopsy. Even today, the reliability of the diagnosis of melanoma can be improved with the support of latest-generation automated analysis tools. Although the human factor continues to be the main discriminating element for the diagnostic accuracy [2], on the other hand such tools could play a relevant role to support the specialist in the diagnosis procedure.
Therefore if, on one hand, it is evident the need to design sophisticated tools supporting the specialists in providing early and accurate diagnosis, on the other hand it is crucial also the necessity to develop low cost solutions alerting in case of suspicious skin lesions. Technology is driving a huge innovation in the healthcare domain and artificial intelligence is playing a key role in providing tools and applications supporting physicians in diagnosis and patient treatment. In particular, computer vision systems are revealing to be fundamental in medical image analysis, since they allow to have non-invasive diagnoses. The specific frameworks for the automatic analysis of skin lesions are constituted by the following fundamental steps: image acquisition, image processing and analysis, features extraction and classification.
This paper is an extension of work originally presented at the 9th
International Conference on Information, Intelligence, Systems and
Applications (IISA 2018). The goal of the original paper [6] was to offer insights into the state of the art in the field of melanoma image classification by both providing a review of the various features used in the elaboration of computer vision systems and focusing on those ones which appear to be the mostly used on the basis of the recent literature. Here a preliminary comparison between two different approaches (Support Vector Machine and Multiple Instance Learning) is proposed, pointing out the key role played by the feature selection (color and texture). In particular, this study was inspired by the good results obtained in [7], where the authors applied Multiple Instance Learning techniques for classifying some medical dermoscopic images drawn from the Ph2 database [8], by using only color features. Our numerical experimentations, reported in this paper, showed that using only color features is not at all high-performance when the classification process is performed on a data base constituted by common plain photographies. On the other hand using in addition texture features provides reasonable classification results, especially if we consider that no pre-processing techniques were used.
This paper is organized as follows. In Section 2 we present the pathogenic mechanisms of the skin cancer, while in Section 3 we recall the main steps adopted in biomedical image processing. In Section 4, a preliminary numerical study is performed for classifying a set of 200 plain photographies (100 melanomas and 100 common nevi), using color and color/texture features. Finally some conclusions are drawn in Section 5, looking at possible future developments.
2. Skin lesions
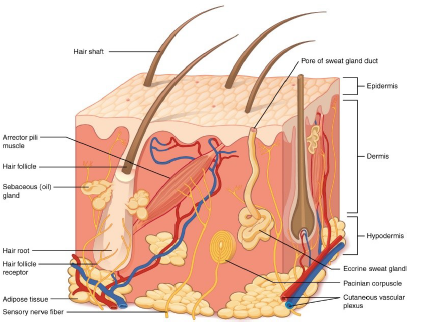
The first layer of the skin is the epidermis, which is constituted by different types of cells, each of them characterized by specific biological chemical functions: keratinocytes have a mechanical function of protection from the external environment, basal cells have a reproductional function of the cells of the most superficial layer and melanocytes provide protection against ultraviolet rays throught the production of melanin.
Dermis is the second layer of the skin, and is constituted by dermal papillary and reticular dermis. It is located below the epidermis and has a thicker consistency (Figure 1). The third layer of the skin is the subcutaneous layer or hypodermis, and, as the dermis, contains many blood and lymphatic vessels and protects the underlying tissues and the internal organs [9].
The different biological structure of the various layers of the skin implies their specific optical properties. When a beam of white light hits the skin, one part is reflected, another part penetrates into the superficial layers and the last part is absorbed. The specific amount of light that is reflected or absorbed depends on genetic factors of the population including the specific phototype to which the individual belongs. Exposure to light triggers melanocytes that produce melanin which is activated as a particularly effective protective filter on shades of blue light.
Normally the melanin deposits are placed in the epidermis (nevi); in the case of malignant lesions, such as the feared melanoma, melanocytes produce excess melanin and these deposits tend to invade the underlying levels of the skin. The presence of melanin in the dermis is one of the most indicative factors in the presence of melanoma (Figure 2).
Further factors indicative of a malignant lesion are the increased blood flow at the periphery of the lesion, evidence of epithelial desquation and the low flow of blood within the lesion which appears with a bluish red hue. Precisely the color features provide clinical evidence for a suspicion of skin cancer. At this point the specialist prescribes a histology to make an explicit diagnosis. Figure 3 shows a common nevus (a), a dysplastic nevus (b) (which is a benign lesion) and finally a melanoma (c) [8].
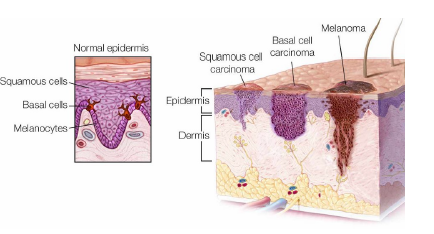 Figure 2: Types of skin cancer
Figure 2: Types of skin cancer
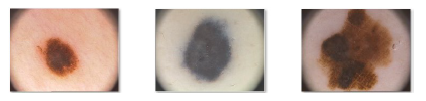 Figure 3: Dermoscopic images of common nevus (a), dysplastic nevus (b) and melanoma (c)
Figure 3: Dermoscopic images of common nevus (a), dysplastic nevus (b) and melanoma (c)
3. Main steps in computer vision systems
In this section we will focus on the fundamental phases that characterize the automatic frameworks for image analysis. We will also refer to the most widespread techniques that characterize the various steps of an artificial vision system: acquisition, pre-processing, segmentation, extraction and selection of features (Figure 4). The classification would be the final step, which is not the main object of this document, apart from some brief discussion in the numerical section.
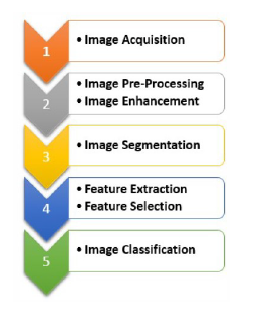 Figure 4: Main steps in biomedical image processing
Figure 4: Main steps in biomedical image processing
3.1 Image acquisition
The first step of computer vision systems is the acquisition of the digital image. The proliferation of techniques as well as the development of new software paradigms has found its way into the numerous techniques that have been presented by researchers and industrial stakeholders. Based on the corresponding pros and cons, each techniques results more suitable for specific cases and less for other ones. Among the mainly widespread techniques we report total cutaneous photography, dermoscopy, ultrasound, magnetic resonance imaging (MRI), confocal scanning laser microscopy (CSLM), positron emission tomography (PET), optical coherence tomography (OCT), multispectral imaging, computed tomography (CT), multifrequency electrical impedance and Raman spectra. The epiluminescence microscopy (ELM), also known as dermoscopy, allows an in-depth analysis of the surface texture of skin lesions as the technique makes the epidermis translucent. Transmission electron microscopy (TEM) is generally used to monitor the evolution of melanoma. Through an appropriate light source the layers of the skin become translucent and this makes possible to appreciate the vascularization of the lesion also analyzing the colorimetric variations of the pigmentation. Computed tomography (CT) is also used to assess the patient’s subjective response, which also allows diagnosis to be made on suspicious lesions.
Positron emission tomography (PET) is a technique that involves the use of contrast fluids through the different absorption of cutaneous melanoma metastases [10]. In the presence of a tumor, the absorption of fluoro-deoxyglucose has a direct correlation with the rate of its proliferation and therefore with the degree of its malignancy. Alternative screening methods are the multifrequency electrical impedance [11] and the Raman spectra [12].
The biological characteristics of a healthy tissue are different from those ones of a diseased tissue. Using the Raman spectra, this different way of diffracting the light coming from a laser beam is exploited, obtaining useful information on the physical structure of the lesion. The spread of wearable devices as well as the enhancement of the smatphones cameras create the conditions for being able to realize applications for auto-screening potentially integrable in platform solutions, through which the patient can interact with specialists aiming at rapid and punctual diagnoses [13]. The increasing costs of health suggest to Governance bodies the adoption of dematerialized health models that can provide decision and responsibility processes, encouraging predictive and preventive analysis [14].
3.2 Image pre-processing
The presence of potential noise linked both to the instrumentation with which the image was acquired, and to the presence of undesired artefacts such as hair and dermatoscopic gel may imply inaccuracies in the diagnosis phase. The objective of the pre-processing phase is to optimize image quality: this phase has a strategic role and is decisive for the entire classification process. The primary task consists in separating the lesion area from the healthy skin. This operation results to be quite difficult as dermoscopic images often present irregular lighting, frames, gelatinous material and intrinsic skin features that make the identification of a lesion, for example trought edges detection methods, a difficult task. Therefore, anything that could affect the image needs to be localized and then removed, masked or replaced. Many techniques have been used in literature in order to perform this task, like image resizing, masking, removal of artefacts, transformation of images into various color spaces. Another important element that commonly penalizes the analysis of dermatoscopic images is the presence of hairs. Recent literature proposes various approaches to optimize images, which involve the use of morphological operators [15], or the detection of curvilinear structures [16] or envisaging a phase of image restructuration through painting [17].
Unfortunately, most of these techniques often leave behind undesirable blurring, invalidating an effective use of the color image. Zhou et al. proposed an algorithm able to carry out an accurate detection of curvilinear artefacts while preserving the lesion structure [18]. The presented approach is very effective for removing the dermatoscopic misuring stick and the hair, but on the other hand it requires high computational requirements. The pre-processing phase of an image can require several operations such as: enhancement, restoration and hair removal (Figure 5).
 Figure 5: Fundamental steps and techniques for pre-processing
Figure 5: Fundamental steps and techniques for pre-processing
Among the various tools to remove unwanted artefacts, the filters are the most immediate ones. Several Computer Vision Systems dedicated to dermatology adopt filters by applying them directly to grayscale photos or on individual color channels in the case of color images [19, 20, 21].
Generally, for each type of unwanted artefacts, a specific strategy involving the use of dedicated solutions is adopted. Many effective methods use color space transformation to optimize both the quantization of color and of contrast, passing from RGB images to different color spaces for enhancing the variation of the color on the edges and inside the lesion.
The pre-processing phase follows antithetical goals. On the one hand, in order to contain the computational complexity, the objective is to identify a minimal number of colors to be used in image analysis. On the other hand, insufficient contrast would make edge detection very difficult. A solution adopted in many circumstances involves the use of histograms, with which it is possible to perform stretching operations to expand the pixel values in the interval [0, 255], or equalization to obtain a uniform distribution of the pixel values. The important task of appropriately enhancing the contrast between the image area where the lesion is contained and the background consisting of healthy skin can be obtained by compensating for irregular lighting through the use of monomorphic filters and high-pass filters. Adaptive and recursive weighted median filter are used to remove air bubbles and dermoscopic gels [22]. An interesting solution involves local processing carried out on a portion of pixels (generally 3×3), assigning the median to the central pixel. Using these techniques, it is possible to eliminate the noise by containing the alterations that may arise in the original image. The strategies referred to, as well as the other solutions presented in the literature, have as their final objective that of being able to favor the segmentation of the image and the extraction of functional features to the classification process. The strategies to be adopted depend on the nature of the considered images and of the impurities that must be managed [23].
3.3 Image segmentation
The analysis and identification of a lesion can be performed directly by specialists who rely on their experience or by using software solutions for image segmentation, such as IMAGEJ or EDISON (Edge Detection and Image Segmentation system). In fact, in melanoma detection, the main task of the segmentation process is to separate the skin lesion from the background consisting of healthy skin (Figure 6), in order to analyze color, structure, shape, texture and information related to vascularization of the lesion.
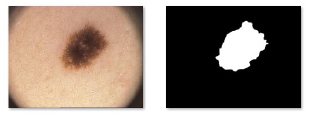 Figure 6: (a) dermatoscopic image (b) segmented lesion
Figure 6: (a) dermatoscopic image (b) segmented lesion
The choice of the color space in which it is best to represent images is important, as it determines the rule of the various color channels. Consequently, it will be important to choose the appropriate segmentation algorithm that takes into account the modes of representation chosen for the images. Without the pretension to be exhaustive, some types of segmentation methods in this field are the following:
- threshold-based, including the Otsu method, those that use the maximum value of the entropy and those that use histograms, such as the independent histogram search algorithm (IHP) [24];
- region-based, including region growing and watershed segmentation;
- pixel-based (aimed at labeling the pixel), both supervised methods such as the Support Vector Machine, unsupervised methods such as fuzzy c-means, and those with reinforcement such as artificial neural networks;
- methods based on customized models, both adaptive and representable through deep multilevel networks.
3.4 Feature extraction
The goal of the segmentation process is to identify the region of the image that contains the lesion by discarding the pixels which are part of the background consisting of healthy skin. In this way the extraction and selection of the most appropriate features to be used for the melanoma detection by artificial vision systems will be favored. In particular, feature extraction implies the enhancement of some specific measures on portions of images: this fundamental step is not often sufficiently discussed and detailed in many researches. On the other hand, a recent study of the relevant features for artificial vision systems in melanoma detection can be found in [6]. As for the extraction of features, many different approaches have been proposed of the statistical and/or filtering type. Among the adopted methods to be mentioned, we cite the principal component analysis, useful for determining the geometric and the shape features, the co-occurrenze matrix [25], useful for the evaluation of texture features, the Fourier spectrum [18], the Gaussian kernels [26], and the transformation of the wavelet package [27], useful for performing image optimization and the extraction of boundaries aimed at reducing data redundancy [28]. In addition, many automated decision support systems implement the feature extraction by exploiting the traditional diagnostic analysis protocols such as the ABCDE system [25], the 7-point checklist [29], the 3-point checklist [30], the model analysis [31] and the Menzies method [32].
4. Numerical experimentations
In this section a preliminary numerical comparison between two different approaches (Support Vector Machine and Multiple Instance Learning) is reported, using color and color/texture features.
|
Table 1: 5-fold cross-validation
Table 2: 10-fold cross-validation
Table 3: Leave-One-Out validation
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
This study starts from some considerations related to the works [7, 33, 34]. In [33] the authors analyzed the role played by the color and the texture features, showing empirically that using only the color features outperforms the use of the texture features: very good results were obtained by means of different type of classifiers on a image data set drawn from the Ph2 database [8], containing 200 melanocytic lesions images (80 common nevi, 80 atypical nevi and 40 melanomas). All the images of the database were selected on the basis of their quality, resolution and dermoscopic features. Good results were obtained also in [7] on the same data set Ph2, by applying Multiple Instance Learning techniques and using only color features.
On the other hand, the objective in [34] was to select the most important image features usable in melanoma detection. In this case, differently from [33] where a high quality data set was adopted, the authors performed their experimentations on a data set constituted by plain photographs publicly available from two online databases https://www.dermquest.coml and http://www.dermins.netl, obtaining very good results in terms of sensitivity (96.77%) and, consequently, in terms of F-score (90.91%). The key point of this work consisted in a very accurate pre-processing and segmentation steps.
On the contrary, the objective of our experimentation consists in simply evaluating the classification results, obtainable on 200 images (100 melanomas and 100 common nevi) drawn from the same database used in [34], but without performing any pre-processing step aimed at cleaning up the images from possible noises [23, 35]. This choice is motivated by the necessity to investigate on the possibility to create fast self-diagnosis systems for accessible skin lesions.
In particular, our experiments were performed by using three configurations of the data set, corresponding respectively to the 5-fold cross-validation, the 10-fold cross-validation and the leaveone-out validation.
For each configuration, initially only the color (RGB) features were used, adding, only successively, those ones related to the texture by means of the co-occurence matrix.
The following classification methods, each of them implemented in Matlab (version R2017b), were adopted:
- Support Vector Machine with linear kernel (SVM)[36]; • Support Vector Machine with RBF kernel (SVM-RBF) [36];
- MIL-RL [37].
Some words are to be spent about the MIL-RL method, proposed in [37]. Such a method belongs the class of the Multiple Instance Learning (MIL) algorithms. The MIL paradigm (see for example
[38, 39]) is a relatively new approach for classification problems which fits very well on image classification, as shown in the recent works [40, 41] and, in particular, also for medical images and video analysis [42]. It differs from the classical supervised classification approaches, since it is aimed at classifying sets of items: such sets are called bags and the items inside the sets are called instances. The main characteristic of a MIL algorithm is that, in the learning phase, only the labels of the bags are known, while the labels of the instances inside the bags are unknown. The MIL paradigm applies very well to image classification since in general an image (bag) is classified on the basis of some portions (instances) of it.
About the MIL-RL code, the same Matlab implementation adopted in [7] were used, while, for the SVM approaches with both linear and RBF kernels, the fitcsvm and predict subroutines for training the classifier and for computing the testing correctness, respectively, were utilized. Such subroutines are included in the Matlab optimization package.
The results of our experimentations are reported in Tables 1, 2 and 3, in terms of accuracy (testing correctness), sensitivity, specificity, F-score and CPU time. For each of these parameters and for each table, the best result was underlined. It is worth noting that the sensitivity is in general more important than the specificity because it measures the capability to identify positive patients. This is in fact taken into account in the F-score value.
Looking at the tables, it is possible to observe that using both color and texture features improves the results, with respect to using only color features. Moreover the better results are generally obtained in corresponding to the MIL-RL algorithm, whose F-score values are however comparable with those ones obtained by using the SVM with RBF kernel. On the other hand using the SVM-RBF is really faster than using the SVM with linear kernel or the MIL-RL approach.
5. Conclusions
In this work some numerical experimentations were performed to classify a data set constituted by plain photos of different sizes, some of which are low resolution images and others are blurred or with hairs. Although in [34] it has been shown that using some pre-processing methodologies on the images improves the classification performances, in our experiments we preferred do not use such techniques. In this way it was intended to operate in the worst conditions in order to compare the classification performances obtainable by using, on one hand, only color features (as proposed in [40, 41, 43]) and, on the other hand, color and texture features. The fact that the used data set consists of plain photographs, instead of high-quality dermatoscopic images such as those ones constituting the Ph2 database, justifies the obtained classification performance.
Adopting additional sets of features together with more sophisticated algorithms providing nonlinear separation surfaces, such as in [44], opens up new possible scenarios for the implementation of efficient solutions, useful to provide a further support to specialists and, on the other hand, to have preliminary self diagnoses, since the diffusion of smartphones allows the possibility to take photos of one’s own skin injuries. The use of medical solutions [45], as well as the possibility of creating Apps, BOTs and Educational Games [46], designed for different classes of population, are possible scenarios of interest.
- R. L. Siegel, K. D. Miller, S. A. Fedewa, D. J. Ahnen, R. G. Meester, A. Barzi, and A. Jemal, “Colorectal cancer statistics, 2017,” CA: a cancer journal for clinicians, vol. 67, no. 3, pp. 177–193, 2017.
- R. Pariser and D. Pariser, “Primary care physicians’ errors in handling cuta- neous disorders: A prospective survey,” Journal of the American Academy of Dermatology, vol. 17, no. 2, pp. 239–245, 1987.
- R. Sanghera and P. S. Grewal, “Dermatological symptom assessment,” in Patient Assessment in Clinical Pharmacy, pp. 133–154, Springer, 2019.
- G. Argenziano, G. Fabbrocini, P. Carli, V. De Giorgi, E. Sammarco, and M. Delfino, “Epiluminescence microscopy for the diagnosis of doubtful melanocytic skin lesions: Comparison of the abcd rule of dermatoscopy and a new 7-point checklist based on pattern analysis,” Archives of Dermatology, vol. 134, no. 12, pp. 1563–1570, 1998.
- S. Menzies, C. Ingvar, and W. McCarthy, “A sensitivity and specificity anal- ysis of the surface microscopy features of invasive melanoma,” Melanoma Research, vol. 6, no. 1, pp. 55–62, 1996.
- E. Vocaturo, E. Zumpano, and P. Veltri, “Features for melanoma lesions characterization in computer vision systems,” in 2018 9th International Con- ference on Information, Intelligence, Systems and Applications, IISA 2018, 2019.
- A. Astorino, A. Fuduli, P. Veltri, and E. Vocaturo, “Melanoma detection by means of multiple instance learning,” Interdisciplinary Sciences: Computa- tional Life Sciences, DOI: 10.1007/s12539-019-00341-y, 2019.
- T. Mendonc¸a, P. M. Ferreira, J. S. Marques, A. R. S. Marcal, and J. Rozeira, “Ph2 – A dermoscopic image database for research and benchmarking,” in 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 5437–5440, 2013.
- G. Anastasi, S. Capitani, M. Carnazza, S. Cinti, O. Cremona, R. De Caro, R. Donato, V. Ferrario, L. Fonzi, A. Franzi, et al., “Trattato di anatomia umana,” 2010.
- C. Pleiss, J. Risse, H.-J. Biersack, and H. Bender, “Role of fdg-pet in the assessment of survival prognosis in melanoma,” Cancer Biotherapy and Ra- diopharmaceuticals, vol. 22, no. 6, pp. 740–747, 2007.
- P. ÅAberg, I. Nicander, J. Hansson, P. Geladi, U. Holmgren, and S. Ollmar, “Skin cancer identification using multifrequency electrical impedance – a po- tential screening tool,” IEEE Transactions on Biomedical Engineering, vol. 51, no. 12, pp. 2097–2102, 2004.
- X. Feng, A. Moy, H. Nguyen, Y. Zhang, M. Fox, K. Sebastian, J. Reichenberg, M. Markey, and J. Tunnell, “Raman spectroscopy reveals biophysical markers in skin cancer surgical margins,” vol. 10490, 2018.
- T. Maier, D. Kulichova, K. Schotten, R. Astrid, T. Ruzicka, C. Berking, and A. Udrea, “Accuracy of a smartphone application using fractal image analysis of pigmented moles compared to clinical diagnosis and histological result,” Journal of the European Academy of Dermatology and Venereology, vol. 29, no. 4, pp. 663–667, 2015.
- E. Vocaturo and P. Veltri, “On the use of networks in biomedicine,” vol. 110, pp. 498–503, 2017.
- P. Schmid, “Segmentation of digitized dermatoscopic images by two- dimensional color clustering,” IEEE Transactions on Medical Imaging, vol. 18, no. 2, pp. 164–171, 1999.
- M. Fleming, C. Steger, J. Zhang, J. Gao, A. Cognetta, L. Pollak, and C. Dyer, “Techniques for a structural analysis of dermatoscopic imagery,” Computerized Medical Imaging and Graphics, vol. 22, no. 5, pp. 375–389, 1998.
- P. Wighton, T. Lee, and M. Atkins, “Dermascopic hair disocclusion using inpainting,” vol. 6914, 2008.
- H. Zhou, G. Schaefer, A. Sadka, and M. Celebi, “Anisotropic mean shift based fuzzy c-means segmentation of dermoscopy images,” IEEE Journal on Selected Topics in Signal Processing, vol. 3, no. 1, pp. 26–34, 2009.
- H. W. Lim, H. Honigsmann, and J. L. Hawk, Photodermatology. CRC Press,2007.
- I. Maglogiannis, S. Pavlopoulos, and D. Koutsouris, “An integrated computer supported acquisition, handling, and characterization system for pigmented skin lesions in dermatological images,” IEEE Transactions on Information Technology in Biomedicine, vol. 9, no. 1, pp. 86–98, 2005.
- M. Anantha, R. Moss, and W. Stoecker, “Detection of pigment network in dermatoscopy images using texture analysis,” Computerized Medical Imaging and Graphics, vol. 28, no. 5, pp. 225–234, 2004.
- A. Dehghani Tafti and E. Mirsadeghi, “A novel adaptive recursive median filter in image noise reduction based on using the entropy,” pp. 520–523, 2013.
- E. Vocaturo, E. Zumpano, and P. Veltri, “Image pre-processing in computer vision systems for melanoma detection,” in IEEE International Conference on Bioinformatics and Biomedicine, BIBM 2018, Madrid, Spain, December 3-6, 2018, pp. 2117–2124, 2018.
- D. Go´mez, C. Butakoff, B. Ersbøll, and W. Stoecker, “Independent histogram pursuit for segmentation of skin lesions,” IEEE Transactions on Biomedical Engineering, vol. 55, no. 1, pp. 157–161, 2008.
- N. Abbasi, H. Shaw, D. Rigel, R. Friedman, W. McCarthy, I. Osman, A. Kopf, and D. Polsky, “Early diagnosis of cutaneous melanoma: Revisiting the abcd criteria,” Journal of the American Medical Association, vol. 292, no. 22, pp. 2771–2776, 2004.
- G. Suro´wka and K. Grzesiak-Kopec´, “Different learning paradigms for the classification of melanoid skin lesions using wavelets,” in Annual Interna- tional Conference of the IEEE Engineering in Medicine and Biology – Pro- ceedings, pp. 3136–3139, 2007.
- C. Lee and D. Landgrebe, “Decision boundary feature extraction for neural networks,” IEEE Transactions on Neural Networks, vol. 8, no. 1, pp. 75–83, 1997.
- I. Maglogiannis, E. Zafiropoulos, and C. Kyranoudis, “Intelligent segmenta- tion and classification of pigmented skin lesions in dermatological images,” Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol. 3955 LNAI, pp. 214–223, 2006.
- M. Healsmith, J. Bourke, J. Osborne, and R. Graham-Brown, “An evaluation of the revised seven-point checklist for the early diagnosis of cutaneous malig- nant melanoma,” British Journal of Dermatology, vol. 130, no. 1, pp. 48–50, 1994.
- H. Soyer, G. Argenziano, I. Zalaudek, R. Corona, F. Sera, R. Talamini, F. Bar- bato, A. Baroni, L. Cicale, A. Di Stefani, P. Farro, L. Rossiello, E. Ruocco, and
S. Chimenti, “Three-point checklist of dermoscopy: A new screening method for early detection of melanoma,” Dermatology, vol. 208, no. 1, pp. 27–31, 2004. - A. Steiner, H. Pehamberger, and K. Wolff, “In vivo epiluminescence mi- croscopy of pigmented skin lesions. ii. diagnosis of small pigmented skin lesions and early detection of malignant melanoma,” Journal of the American Academy of Dermatology, vol. 17, no. 4, pp. 584–591, 1987.
- S. W. Menzies, K. A. Crotty, C. Ingvar, and W. H. McCarthy, An atlas of surface microscopy of pigmented skin lesions: dermoscopy. McGraw-Hill Sydney, Australia, 2003.
- C. Barata, M. Ruela, M. Francisco, T. Mendonca, and J. Marques, “Two systems for the detection of melanomas in dermoscopy images using texture and color features,” IEEE Systems Journal, vol. 8, no. 3, pp. 965–979, 2014.
- S. Mustafa and A. Kimura, “A svm-based diagnosis of melanoma using only useful image features,” in 2018 International Workshop on Advanced Image Technology, IWAIT 2018, pp. 1–4, 2018.
- E. Vocaturo, E. Zumpano, and P. Veltri, “On the usefulness of pre-processing step in melanoma detection using multiple instance learning,” in 13th Inter- national Conference on Flexible Query Answering Systems (FQAS) 2019, in press, 2019.
- V. Vapnik, The nature of statistical learning theory. Springer science & business media, 2013.
- A. Astorino, A. Fuduli, and M. Gaudioso, “A Lagrangian relaxation approach for binary multiple instance classification,” IEEE Transactions on Neural Networks and Learning Systems, vol. 30, pp. 2662–2671, 2019.
- J. Amores, “Multiple instance classification: review, taxonomy and compara- tive study,” Artificial Intelligence, vol. 201, pp. 81–105, 2013.
- M.-A. Carbonneau, V. Cheplygina, E. Granger, and G. Gagnon, “Multiple in- stance learning: A survey of problem characteristics and applications,” Pattern Recognition, vol. 77, pp. 329–353, 2018.
- A. Astorino, A. Fuduli, M. Gaudioso, and E. Vocaturo, “A multiple instance learning algorithm for color images classification,” in Proceedings of the 2Nd International Database Engineering & Applications Symposium, IDEAS 2018, (New York, NY, USA), pp. 262–266, ACM, 2018.
- A. Astorino, A. Fuduli, M. Gaudioso, and E. Vocaturo, “Multiple instance learning algorithm for medical image classification,” in Proceedings of the 27th Italian Symposium on Advanced Database Systems, Castiglione della Pescaia (Grosseto), Italy, June 16-19, 2019, 2019.
- G. Quellec, G. Cazuguel, B. Cochener, and M. Lamard, “Multiple-instance learning for medical image and video analysis,” IEEE Reviews in Biomedical Engineering, vol. 10, pp. 213–234, 2017.
- A. Astorino, A. Fuduli, P. Veltri, and E. Vocaturo, “On a recent algorithm for multiple instance learning. preliminary applications in image classification,” in Proceedings – 2017 IEEE International Conference on Bioinformatics and Biomedicine, BIBM 2017, vol. 2017-January, pp. 1615–1619, 2017.
- M. Gaudioso, G. Giallombardo, G. Miglionico, and E. Vocaturo, “Classifi- cation in the multiple instance learning framework via spherical separation,” Soft Computing, DOI: 10.1007/s00500-019-04255-1, in press, 2019.
- E. Zumpano, P. Iaquinta, L. Caroprese, G. Cascini, F. Dattola, P. Franco,M. Iusi, P. Veltri, and E. Vocaturo, “Simpatico 3d: A medical information system for diagnostic procedures,” in IEEE International Conference on Bioinformatics and Biomedicine, BIBM 2018, Madrid, Spain, December 3-6, pp. 2125–2128.
- E. Vocaturo, E. Zumpano, L. Caroprese, S. M. Pagliuso, and D. Lappano, “Educational games for cultural heritage,” in Proceedings of 1st International Workshop on Visual Pattern Extraction and Recognition for Cultural Her- itage Understanding co-located with 15th Italian Research Conference on Digital Libraries (IRCDL 2019), CNR Area in Pisa, Italy, January 30, 2019.,pp. 95–106, 2019.