Efficient Alignment of Very Long Sequences
Efficient Alignment of Very Long Sequences
Volume 3, Issue 2, Page No 329-345, 2018
Author’s Name: Chunchun Zhaoa), Sartaj Sahni
View Affiliations
Department of Computer and Information Science and Engineering, University of Florida, Gainesville, FL, USA
a)Author to whom correspondence should be addressed. E-mail: czhao@cise.ufl.edu
Adv. Sci. Technol. Eng. Syst. J. 3(2), 329-345 (2018); ![]() DOI: 10.25046/aj030236
DOI: 10.25046/aj030236
Keywords: Sequence alignment, Energy efficient, Multi core
Export Citations
We consider the problem of aligning two very long biological sequences. The score for the best alignment may be found using the Smith-Waterman scoring algorithm while the best alignment itself may be determined using Myers and Miller’s alignment algorithm. Neither of these algorithms takes advantage of computer caches to obtain high efficiency. We propose cache-efficient algorithms to determine the score of the best alignment as well as the best alignment itself. All algorithms were implemented using C and OpenMP, and benchmarked using real data sets from the National Center for Biotechnology Information (NCBI) database. The test computational platforms were Xeon E5 2603, I7-x980 and Xeon E5 2695. Our best single-core cacheefficient scoring algorithm reduces the running time by as much as 19.7% relative to the Smith-Waterman scoring algorithm and our best cache-efficient alignment algorithm reduces the running time by as much as 17.1% relative to the Myers and Miller alignment algorithm. Multicore versions of our cache-efficient algorithms scale quite well up to the 24 cores we tested; achieving a speedup of 22 with 24 cores. Our multi-core scoring and alignment algorithms reduce the running time by as much as 61.4% and 47.3% relative to multi-core versions of the Smith-Waterman scoring algorithm and Myers and Miller’s alignment algorithm, respectively.
Received: 20 March 2018, Accepted: 16 April 2018, Published Online: 30 April 2018
1. Introduction
Sequence alignment is a fundamental and well-studied problem in the biological sciences. In this problem, we are given two sequences A[1 : m] = aa2···am and B[1 : n] = b1b2···bn and we are required to find the score of the best alignment and possibly also an alignment with this best score. When aligning two sequences, we may insert gaps into the sequences. The score of an alignment is determined using a matching (or scoring) matrix that assigns a score to each pair of characters from the alphabet in use as well as a gap penalty model that determines the penalty associated with a gap sequence. In the linear gap penalty model, the penalty for a gap sequence of length k > 0 is kg, where g is some constant while in the affine model this penalty is gopen + (k − 1) ∗ gext. The affine model more accurately reflects the fact that opening a gap is more expensive than extending one. Two versions of sequence alignment–global and local–are of interest. In global alignment, the entire A sequence is to be aligned with alignment score. The alphabet for DNA, RNA, and protein sequences is, respectively, {A, T, G, C}, {A, U, G, C}, and {A, C, D, E, F, G, H, I, K, L, M, N, P, Q, R, S, T, V, W, Y}.
Figure 1 illustrates these concepts using the DNA sequences A[1 : 8] = {AGTACGCA} and B[1 : 5] = {TATGC}. The symbol ‘_’ denotes the gap character. The alignment of Figure 1(a) is a global alignment and that of Figure 1(b) is a local one. To score the alignments, we have used the linear penalty model with g = −2 and the scores for pairs of aligned characters, which are taken from BLOSUM62 matrix in [1], are c(T,T) = 5, c(A,A) = 4, c(C,C) = 9, c(G,G) = 6, and c(C,T) = −1. The score for the shown global alignment is 17 while that for the shown local alignment is 23. If we were using an affine penalty model with gopen = −4 and gext = −2, then the penalty for each of the gaps in positions 1 and 8 of the global alignment would be −4 and the overall score for the global alignment would be 13.
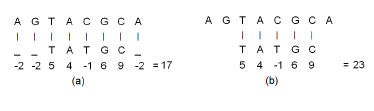 Figure 1: Example alignments using the linear gap penalty model. (a) Global alignment (b) Local alignment
Figure 1: Example alignments using the linear gap penalty model. (a) Global alignment (b) Local alignment
In [2], the authors first proposed an O(mn) time algorithm, called Needleman-Wunsch(NW) algorithm, for global alignment using the linear gap model. This algorithm requires O(n) space when only the score of the best alignment is to be determined and O(mn) space when the best alignment is also to be determined. In [3], the authors proposed a new algorithm called SmithWaterman(SW) algorithm, which modified the NW algorithm so as to determine the best local alignment. In [4], the author proposed a dynamic programming algorithm called Gotoh algorithm, for sequence alignment using an affine gap penalty model. The asymptotic complexity of the SW and Gotoh algorithms is the same as that of the NW algorithm.
When mn is large and the best alignment is sought, the space, O(mn), required by the algorithms of NW, SW and Gotoh exceeds what is available on most computers. The best alignment for these large instances can be found in [5] using sequence alignment algorithms derived from Hirschberg’s linear space divide-and-conquer algorithm for the longest common subsequence problem. In [6], the authors developed a Myers-Miller alignment algorithm. It is the linear space O(mn) time version of Hirschberg’s algorithm for global sequence alignment using an affine gap penalty model. And in [7], the authors do this for local alignment.
In an effort to speed sequence alignment, fast sequence-alignment heuristics have been developed. As in [8, 9, 10],BLAST, FASTA, and Sim2 are a few examples of software systems that employ sequence alignment heuristics. Another direction of research, also aimed at speeding sequence alignment, has been the development of parallel algorithms. Parallel algorithms for sequence alignment may be found in [11]-[19], for example.
In this paper, we focus on reducing the number of cache misses that occur in the computation of the score of the best alignment as well as in determining the best alignment. Although we explicitly consider only the linear gap penalty model, our methods readily extend to the affine gap penalty model. Our interest in cache misses stems from two observations–(1) the time required to service a last-level-cache (LLC) miss is typically 2 to 3 orders of magnitude more than the time for an arithmetic operation and (2) the energy required to fetch data from main memory is typically between 60 to 600 times that needed when the data is on the chip. As a result of observation (1), cache misses dominate the overall running time of applications for which the hardware/software cache prefetch modules on the target computer are ineffective in predicting future cache misses. The effectiveness of hardware/software cache prefetch mechanisms varies with application, computer, and compiler. So, if we are writing code that is to be used on a variety of computer platforms, it is desirable to write cache-efficient code rather than to rely exclusively on the cache prefetching of the target platform. Even when the hardware/software prefetch mechanism of the target platform is very effective in hiding memory latency, observation (2) implies excessive energy use when there are many cache misses.
This paper is an extension of work originally in [20], which has been presented by us in the 2015 IEEE 5th international conference on Computational Advances
in Bio and Medical Sciences (ICCABS). The main contributions are
- cache efficient single-core and multi-core algorithms to determine the score of the best alignment;
- cache efficient single-core and multi-core algorithms to determine the best alignment.
The rest of the paper is organized in the following way. In Section 2, we describe our cache model. Our cache-efficient algorithms for scoring and alignment are developed and analyzed in Section 3. Experimental results are presented in Section 4. In Section 5, we present a discussion of these results and in Section 6, we present the limitations of our work. Finally, we conclude in Section 7.
2. Cache Model
For simplicity in the analysis, we assume a single cache comprised of s lines of size w words (a word is large enough to hold a piece of data, typically 4 bytes) each. So, the total cache capacity is sw words. The main memory is partitioned into blocks also of size w words each. When the program needs to read a word that is not in the cache, a cache miss occurs. To service this cache miss, the block of main memory that includes the needed word is fetched and stored in a cache line, which is selected using the LRU (least recently used) rule. Until this block of main memory is evicted from this cache line, its words may be read without additional cache misses. We assume the cache is written back with write allocate. Write allocate means that when the program needs to write a word of data, a write miss occurs if the block corresponding to the main memory is not currently in cache. To service the write miss, the corresponding block of main memory is fetched and stored in a cache line. Write back means that the word is written to the appropriate cache line only. A cache line with changed content is written back to the main memory when it is about to be overwritten by a new block from main memory.
Rather than directly assess the number of read and write misses incurred by an algorithm, we shall count the number of read and write accesses to main memory.
Every read and write miss makes a read access. A read and write miss also makes a write access when the data in the replacement cache line is written to main memory.
We emphasize that the described cache model is a very simplified model. In practice, modern computers commonly have two or three levels of cache and employ sophisticated adaptive cache replacement strategies rather than the LRU strategy described above. Further, hardware and software cache prefetch mechanisms are often deployed to hide the latency involved in servicing a cache miss. These mechanisms may, for example, attempt to learn the memory access pattern of the current application and then predict the future need for blocks of main memory. The predicted blocks are brought into cache before the program actually tries to read/write from/into those blocks thereby avoiding (or reducing) the delay involved in servicing a cache miss. Actual performance is also influenced by the compiler used and the compiler options in effect at the time of compilation. As a result, actual performance may bear little relationship to the analytical results obtained for our simple cache model. Despite this, we believe the simple cache model serves a useful purpose in directing the quest for cache-efficient algorithms that eventually need to be validated experimentally.
3. Cache Efficient Algorithms
3.1. Scoring Algorithms
3.1.1 Needleman-Wunsch and Smith-Waterman algorithm
Let Hij be the score of the best global alignment for
A[1 : i] and B[1 : j]. We wish to determine Hmn. In [2], the authors derived the following dynamic programming equations for H using the linear gap penalty model. These equations may be used to compute Hmn.
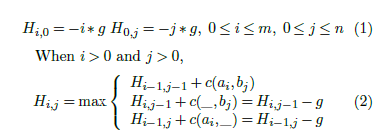 Hi−1,j +c(ai,_) = Hi−1,j −g where c(ai,bj) is the match score between characters ai and bj and g is the gap penalty.
Hi−1,j +c(ai,_) = Hi−1,j −g where c(ai,bj) is the match score between characters ai and bj and g is the gap penalty.
For local alignment, Hij denotes the score of the best local alignment for A[1 : i] and B[1 : j]. In [3], the Smith-Waterman equations for local alignment using the linear gap penalty model are:
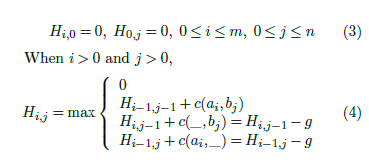 Several authors (in [5, 6], for example) have observed that the score of the best local alignment may be determined using a single array H[0 : n] as in algorithm Score(Algorithm 1.)
Several authors (in [5, 6], for example) have observed that the score of the best local alignment may be determined using a single array H[0 : n] as in algorithm Score(Algorithm 1.)
Algorithm 1 Smith-Waterman scoring algorithm
1: Score(A[1 : m],B[1 : n]) 2: for j ← 0 to n do
3: H[j] ← 0 //Initialize row 0
4: end for 5: for i ← 1 to m do
6: diag ← 0 // Compute row i
7: for j ← 1 to n do
8: nextdiag ← H[j]
9: H[j] ← max{0,diag + c(A[i],B[j]),H[j − 1] − g,H[j]−g}
10: diag ← nextdiag
11: end for
12: end for
13: return H[n]
The scoring algorithm for the Needleman and Wunsch algorithm is similar. It is easy to see that the time complexity of the algorithm of Algorithm 1 is O(mn) and its space complexity is O(n).
 Figure 2: Memory access pattern for Score algorithm(Algorithm 1).
Figure 2: Memory access pattern for Score algorithm(Algorithm 1).
For the (data) cache miss analysis, we focus on read and write misses of the array H and ignore misses due to the reads of the sequences A and B as well as of the scoring matrix c (notice that there are no write misses for A, B, and c). Figure 2 shows the memory access pattern for H by algorithm Score. The first row denotes the initialization of H and subsequent rows denote access for different value of i (i.e., different iterations of the for i loop). The computation of Hij is done using a single one-dimensional array H[], following the i’th iteration of the for i loop, H[j] = Hij. In each iteration of this loop, the elements of H[] are accessed left-to-right. During the initialization loop, H is brought into the cache in blocks of size w. Assume that n is sufficiently large so that H[] does not entirely fit into the cache. Hence, at some value of j, the cache capacity is reached and further progress of the initialization loop causes the least recently used blocks of H[]
(i.e., blocks from left to right) to be evicted from the cache. The evicted blocks are written to main memory as they have been updated. So, the initialization loop results in n/w read accesses and (approximately) n/w write accesses (the number of write accesses is actually n/w −s). Since the left part of H[] has been evicted from the cache by the time we start the computation for row i (i > 0), each iteration of the for i loop also results in n/w read accesses and approximately n/w write accesses. So, the total number of read accesses is (m+1)n/w ≈ mn/w and the number of write accesses is also ≈ mn/w. The number of read and write accesses is ≈ 2mn/w, when n is large.
We note, however, that when n is sufficiently small that H[] fits into the cache, the number of read accesses is n/w (all occur in the initialization loop) and there are no write accesses. In practice, especially in the case of local alignment involving a very long sequence, one of the two sequences A and B is small enough to fit in the cache while the other may not fit in the cache. So, in these cases, it is desirable to ensure that A is the longer sequence and B is the shorter one so that H fits in the cache entirely. This is accomplished by swap A and B sequences.
When m < n and H[1 : m] fits into the cache and
H[1 : n] does not, algorithm Score incurs O(mn/w) read/write accesses, while swap A and B incurs O(m/w) read/write accesses.
3.1.2. Diagonal Algorithm
An alternative to computing the score by rows is to compute by diagonals. While this uses two one-dimensional arrays rather than one, it is more readily parallelized than Score as all elements on a diagonal can be computed at the same time; elements on a row need to be computed in sequence. Algorithm 2 Diagonal scoring algorithm
1: Diagonal(A[1 : m],B[1 : n])
2: d2[0] ← d1[0] ← d1[1] ← 0
3: for d ← 2 to m+n do
4: x ← (d <= m?0 : d−m);y ← (d <= n?d : n); for i ← x to y do
j ← d−i
diag ← next+c(A[i]+B[j]) left ← d1[i−1]−g up ← d1[i]−g next ← d2[i]
11: d2[i] ← max{0,diag,left,up}
12: end for
13: swap(d1,d2)
14: end for
15: return d1[n]
Algorithm Diagonal (Algorithm 2) uses two onedimensional arrays d1[] and d2[], where d2[] stores the scores for the (d−2)th diagonal data and d1[] stores them for the (d−1)th diagonal. When we compute the element Hi,j in the dth diagonal, the previous diagonal element Hi−1,j−1 is fetched from d2[] and the previous left element Hi−1,j and previous upper element Hi,j−1 are fetched from d1[]. The calculated Hi,j overwrites the old value in d2[].
The total number of read accesses is mn/w for each diagonal array and the total number of write accesses is mn/w for both arrays combined. The number of cache misses for Diagonal is approximately 3mn/w when n is large.
3.1.3. Strip Algorithm
When neither H[1 : m] nor H[1 : n] fits into the cache, accesses to main memory may be reduced by computing Hij by strips of width q such that q consecutive elements of H[] fit into the cache. Specifically, we partition H[1 : n] into n/q strips of size q (except possibly the last strip whose size may be smaller than q) as in Figure 3. First, all Hij in strip 0 are computed, then those in strip 1, and so on. When computing the values in a strip, we need those in the rightmost column of the preceding strip. So, we save these rightmost values in a one-dimensional array strip[0 : m]. The algorithm is given in Algorithm 3. We note that sequence alignment by strips has been considered before. For example, in [12], the authors using the similar approach in their GPU algorithm. Their use differs from ours in that they compute the strips in pipeline fashion with each strip assigned to a different pipeline stage in round robin fashion and within a strip, the computation is done by anti-diagonals in parallel. On the other hand, we do not pipeline the computation among strips and within a strip, our computation is by rows. Algorithm 3 Strip scoring algorithm
1: Strip(A[1 : m],B[1 : n])
2: for j ← 1 to m do
3: strip[j] ← 0 //leftmost strip
4: end for
5: for t ← 1 to n/q do 6: for j ← t∗q to t∗q +q −1 do
7: H[j] ← 0 //Initialize first row
8: end for for i ← 1 to m do
diag ← strip[i−1]
H[t∗q −1] ← strip[i]
for j ← t∗q to t∗q +q −1 do nextdiag ← H[j]
H[j] ← max{0,diag+c(A[i],B[j]),H[j−1]− g,H[j]−g}
15: diag ← nextdiag
16: end for
17: strip[i] ← H[t∗q +q −1]
18: end for
19: end for
20: return H[n]
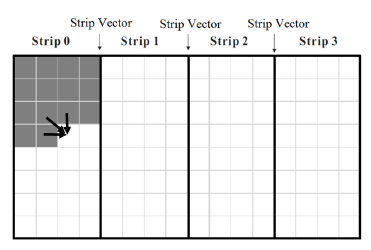 Figure 3: Memory access pattern for Strip algorithm (Algorithm 3).
Figure 3: Memory access pattern for Strip algorithm (Algorithm 3).
It is easy to see that the time complexity of algorithm Strip is O(mn) and that its space complexity is O(m+n). For the cache misses, we focus on those resulting from the reads and writes of H[] and strip[]. The initialization of strip results in m/w read accesses and approximately the same number of write accesses. The computation of each strip makes the following accesses to main memory:
- q/w read accesses for the appropriate set of q entries of H for the current strip and q/w write accesses for the cache lines whose data are replaced by these H The write accesses are, however, not made for the first strip.
- m/w read accesses for strip and m/w write accesses. The number of write accesses is less by s for the last strip.
So, the overall number of read accesses is m/w +
(q/w +m/w)∗n/q = m/w +n/w +mn/(wq) and the number of write accesses is approximately the same as this. So, the total number of main memory accesses is ≈ 2mn/(wq) when m and n are large.
3.2. Alignment Algorithms
In this section, we examine algorithms that compute the alignment that results in the best score rather than just the best score. While in the previous section we explicitly considered local alignment and remarked that the results readily extend to global alignment, in this section we explicitly consider global alignment and remark that the methods extend to local alignment.
3.2.1. Myers and Miller’s Algorithm
When aligning very long sequences, the O(mn) space requirement of the full-matrix algorithm exceeds the available memory on most computers. For these instances, we need a more memory-efficient alignment algorithm. In [6], Myers and Miller have adapted Hirschberg’s linear space algorithm for the longest common subsequence problem to find the best global alignment in linear space. Its time complexity is O(mn). However, this linear space adaptation performs about twice as many operations as does the full-matrix algorithm. In [11], the authors have developed a hybrid algorithm, FastLSA, whose memory requirement adapts to the amount of memory available on the target computing platform. In this section and the next, we focus on the adaptation of Myers-Miller algorithm.
It is easy to see that an optimal (global) alignment is comprised of an optimal alignment of A[1 : m/2] and B[1 : j] and an optimal alignment of A[m : m/2 + 1] (A[m : i] is the reverse of A[i : m]) and B[n : j +1] for some j, 1 ≤ j ≤ n. The value of j for which this is true is called the optimal crossover point. Myers and Miller’s linear space algorithm for alignment determines the optimal alignment by first determining the optimal crossover point (si,sj) where si = m/2, and then recursively aligning A[1 : m/2] and B[1 : sj] as well as A[m : m/2+1] and B[n : sj +1]. Equivalently, an optimal alignment of A[1 : m] and B[1 : n] is an optimal alignment of A[1 : m/2] and B[1 : sj] concatenated with the reverse of an optimal alignment of A[m : m/2+1] and B[n : sj +1]. Hence, an optimal alignment is comprised of a sequence of optimal crossover points. This is depicted visually in Figure 4. Figure 4(a) shows alignments using 3 possible crossover points at row m/2 of H. Figure 4(b) shows the partitioning of the alignment problem into 2 smaller alignment problems (shaded rectangles) using the optimal crossover point (meeting point of the 2 shaded rectangles) at row m/2. Figure 4(c) shows the partitioning of each of the 2 subproblems of Figure 4(b) using the optimal crossover points for these subproblems (note that these crossovers take place at rows m/4 and 3m/4, respectively). Figure 4(d) shows the constructed optimal alignment, which is presently comprised of the 3 determined optimal crossover points.
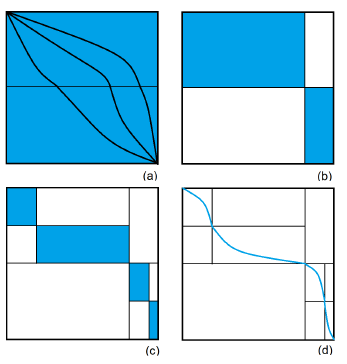 Figure 4: An alignment as crossover points. (a) Three alignments (b) Optimal crossover for m/2 (c)Optimal crossover points at m/4 and 3m/4 (d) Best alignment Algorithm 4 Myers and Miller algorithm
Figure 4: An alignment as crossover points. (a) Three alignments (b) Optimal crossover for m/2 (c)Optimal crossover points at m/4 and 3m/4 (d) Best alignment Algorithm 4 Myers and Miller algorithm
1: MM(A[1 : m],B[1 : n],path[1 : m]) 2: if m <= 1 or n <= 1 then
3: Linear search to find optimal crossover point [si,sj]
4: path[m] ← [si,sj]
5: else
6: Htop ← MScore
7: Hbot ← MScore
8: Linear search to find optimal crossover point [si,sj]
9: MM,path
10: path
11: MM ,path
12: end if
The Myers and Miller algorithm, MM (Algorithm 4), uses a modified version of the linear space scoring algorithm Score (Algorithm 1) to obtain the scores for the best alignments of A[1 : i] and B[1 : j], 1 ≤ i ≤ m/2, 1 ≤ j ≤ n as well as for the best alignments of A[m : i] and B[m : j], m/2 < i ≤ m, 1 ≤ j ≤ n. This modified version MScore differs from Score only in that MScore returns the entire array H rather than just H[n]. Using the returned H arrays for the forward and reverse alignments, the optimal crossover point for the best alignment is computed as in algorithm MM (Algorithm 4). Once the optimal crossover point is known, two recursive calls are made to optimally align the top and bottom halves of A with left and right parts of B. The approximately time complexity for iteration k is O(2mn/2k), hence the total time complexity is rough 2mn.
In each level of recursion, the number of main memory accesses is dominated by those made in the calls to MScore. From the analysis for Score, it follows that when n is large, the number of accesses to main memory is ≈ 2mn/w(1+1/2+1/4+···) ≈ 4mn/w.
3.2.2. Diagonal Myers and Miller Algorithm
Let Mdiagonal be algorithm Diagonal (Algorithm 2) modified to return the entire H array rather than just H[n]. Our diagonal Myers and Miller algorithm (MMDiagonal) replaces the two statements in algorithm MM (Algorithm 4) that invoke MScore with a test that causes Mdiagonal to be used in place of MScore when both of m and n are sufficiently long.
From the analysis for Diagonal, it follows that when m and n are large, the number of accesses to main memory is ≈ 3mn/w(1+1/2+1/4+···) ≈ 6mn/w.
3.2.3. Striped Myers and Miller Algorithm
Let MStrip be algorithm Strip (Algorithm 3) modified to return the entire H array rather than just H[n]. Our striped Myers and Miller algorithm (MMStrip) replaces the two statements in algorithm MM (Algorithm 4) that invoke MScore with a test that causes MStrip to be used in place of MScore when both of m and n are sufficiently long.
From the analysis for Strip, it follows that when m and n are large, the number of accesses to main memory is ≈ 2mn/(wq)(1+1/2+1/4+···) ≈ 4mn/(wq).
3.3. Parallel Scoring Algorithms
3.3.1. Parallel Score Algorithm
As remarked earlier in Score algorithm, the elements in a row of the score matrix need to be computed sequentially from left to right because of data dependencies. So, we are unable to parallelize the inner for loop of Score (Algorithm 1). Instead, we adopt the unusual approach of parallelizing the outer for loop while computing the inner loop sequentially using a single processor. Initially, processor s is assigned to do the outer loop computation for i = s, 1 ≤ i ≤ p, where p is the number of processors. Processor s begins after a suitable time lag relative to the start of processor s−1 so that the data it needs for its computation has already been computed by processor s−1. That is, processor 1 begins the inner loop computation for i = 1 at time 0, then, with a suitable time lag, processor 2 begins the outer loop computation for i = 2, then, with a further lag, processor 3 begins the i = 3 computation and so on. When a processor has finished with its iteration i computation, it starts on iteration i+p of the outer loop. Synchronization primitives are used to ensure suitable time lags. The time complexity of the resulting p-core algorithm PP_Score is O(mn/p).
3.3.2. Parallel Diagonal Algorithm
The inner for loop of Diagonal (Algorithm 2) is easily parallelized as the elements on a diagonal are independent and may be computed simultaneously. So, in our parallel version, we divide the diagonal d into p blocks, where p is the number of processors. We assign a block to each processor from left to right as in Figure 5. The time complexity of the resulting p-core algorithm PP_Diagonal is O(mn/p).
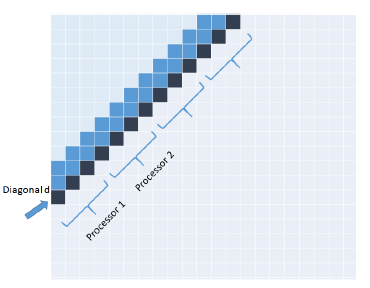 Figure 5: Parallel Diagonal algorithm.
Figure 5: Parallel Diagonal algorithm.
3.3.3. Parallel Strip Algorithm
In the Strip scoring algorithm, we partition the score matrix H into n/q strips of size q (Figure 3) and compute the strips one at a time from left to right. Inside a strip, scores are computed row by row from top to bottom. We see that the computation of one strip can begin once the first row of the previous strip has been computed. In our parallel version of this algorithm, processor i is initially assigned to compute strip i, 1 ≤ i ≤ p. When computing a value in its assigned strip, a processor needs to wait until the values (if any) needed from the strip to its left have been computed. When a processor completes the computation of strip j, it proceeds with the computation of strip j +p. Figure 6 shows a possible state in the described parallel strip computation strategy. We maintain an array signal[] such that signal[r] = s+1 iff the row r computation for strips 1 through s has been completed. This array enables the processor working on the strip to its right to determine when it can begin the computation of its r’th row. The time complexity of the resulting parallel strip algorithm, PP_Strip, is O(mn/p).
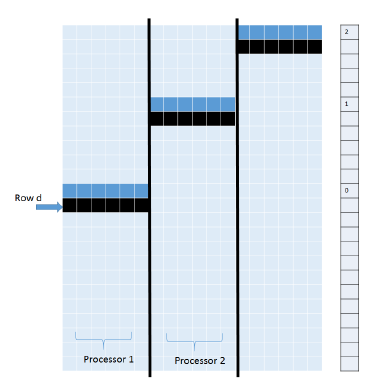 Figure 6: Parallel Strip algorithm.
Figure 6: Parallel Strip algorithm.
3.4. Parallel Alignment Algorithms
In the single-core implementation, we divide the H matrix into two equal size parts and apply the scoring algorithm to each part. Then, we determine the optimal crossover point where the sum of the scores from both directions is maximum. This crossover point is used to divide the matrix into two smaller score matrices to which this decomposition strategy is recursively applied. The first application of this strategy yields two independent subproblems and following an application of the strategy to each of these subproblems, we have 4 even smaller subproblems. Following k rounds, we have 2k independent subproblems.
For the parallel version of alignment algorithms, we employ the following strategies:
- When the number of independent matrices is small, each matrix is computed using the parallel version of score algorithms PP_Score, PP_Diagonal and PP_Strip; where p processors are assigned to the parallel computation. In other words, the matrices are computed in sequence.
- When the number of independent matrices is large, each matrix is computed using the single-core al-
gorithms Score, Diagonal and Strip. Now, p matrices are concurrently computed.
Let PP_MM, PP_MMDiagonal and
PP_MMStrip, respectively, denote the parallel versions of MM, MMDiagonal and MMStrip.
4. Results
4.1. Experimental Settings and Test Data
We implemented the single-core scoring and alignment algorithms in C and the multi-core scoring and alignment algorithms in C and OpenMP. The relative performance of these algorithms was measured on the following platforms:
- Intel Xeon CPU E5-2603 v2 Quad-Core processor
1.8GHz with 10MB cache.
- Intel I7-x980 Six-Core processor 3.33GHz with 12MB LLC cache.
- Intel Xeon CPU E5-2695 v2 2xTwelve-Core processors 2.40GHz with 30MB cache.
For convenience, we will, at times, refer to these platforms as Xeon4, Xeon6, and Xeon24 (i.e., the number of cores is appended to the name Xeon).
All codes were compiled using the gcc compiler with the O2 option. On our Xeon4 platform, we used the “perf” [21] software to measure energy usage through the RAPL interface. So, for this platform, we report cache misses and energy consumption as well as running time. For the Xeon6 and Xeon24 platforms, we provide the running time only.
For test data, we used randomly generated protein sequences as well as real protein sequences obtained from the Globin Gene Server[22] and DNA/RNA/protein sequences from the National Center for Biotechnology Information (NCBI) database [23]. We used the BLOSUM62[1] scoring matrix for all our experiments. The results for our randomly generated protein sequences were comparable to those for similarly sized sequences used from the two databases [22] and [23]. So, we present only the results for the latter data sets here.
4.2. Xeon E5-2603 (Xeon4)
4.2.1. Score Algorithms
Figure 7 and Table 1 give the number of cache misses on our Xeon4 platform for different sequence sizes. The last two columns of Table 1 gives the percent reduction in the observed cache miss count of Strip relative to Score and Diagonal. Strip has the fewest cache misses followed by Score and Diagonal (in this order). Strip reduces cache misses by up to 86.2% relative to Score and by up to 92.3% relative to Diagonal.
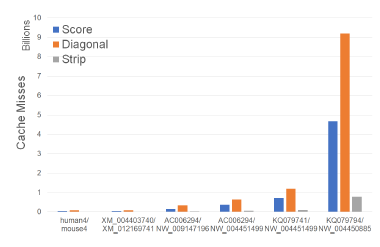 Figure 7: Cache misses of scoring algorithms, in billions, on Xeon4.
Figure 7: Cache misses of scoring algorithms, in billions, on Xeon4.
Figure 8 and Table 2 give the running times of our scoring algorithms on our Xeon4 platform. In the figure, the time is in seconds while in the table, the time is given using the format hh : mm : ss. The table also gives the percent reduction in running time achieved by Strip relative to Score and Diagonal.
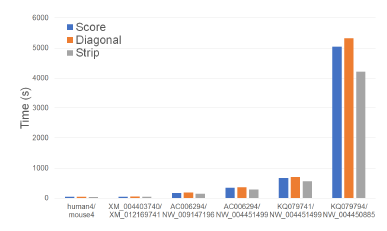 Figure 8: Run time of scoring algorithms, in seconds, on Xeon4.
Figure 8: Run time of scoring algorithms, in seconds, on Xeon4.
As can be seen, on our Xeon4 platform, Strip is the fastest followed by Score and Diagonal (in this order). Strip reduces the running time by up to 17.5% relative to Score and by up to 22.8% relative to Diagonal. The reduction in running time, while significant, isn’t as much as the reduction in cache misses possibly due to the effect of cache prefetching, which reduces cache induced computational delays.
Figures 9 and Tables 3 give the CPU and cache energy consumed, in joules, by our Xeon4 platform.
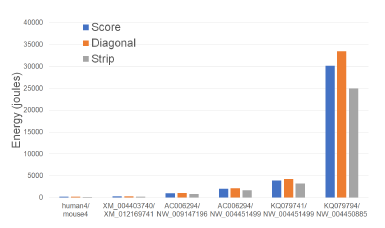 Figure 9: CPU and cache energy consumption of scoring algorithms, in joules, on Xeon4.
Figure 9: CPU and cache energy consumption of scoring algorithms, in joules, on Xeon4.
On our datasets, Strip required up to 18.5% less CPU and cache energy than Score and up to 25.5% less than Diagonal. It is interesting to note that the energy reduction is comparable to the reduction in running time suggesting a close relationship between running time and energy consumption for this application.
4.2.2. Parallel Scoring Algorithms
Figure 10 and Table 4 give the number of cache misses on our Xeon4 platform for our parallel scoring algorithms. PP_Strip has the fewest cache misses followed by PP_Score and PP_Diagonal (in this order). PP_Strip reduces cache misses by up to 98.1% relative to PP_Score and by up to 99.1% relative to PP_Diagonal. We observe also that the total cache misses for PP_Score is slightly higher than for Score for smaller instances and lower for larger instances. PP_Diagonal, on the other hand, consistently has more cache misses than Diagonal. PP_Strip exhibits a significant reduction in cache misses. This is because we chose the strip width to be such that p strip rows fit in this cache. Most of the cache misses in the Strip are from the vector that transfers boundary results from one strip to the next. When p strips are being worked on simultaneously, the inter-strip data that is to be transferred is often in the cache and so many of the cache misses incurred by the single-core algorithm are saved. The remaining two algorithms do not allow this flexibility in choosing the segment size a processor works on; this size is fixed at O(n/p).
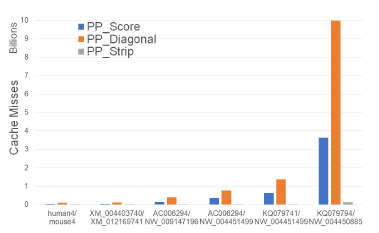 Figure 10: Cache misses of parallel scoring algorithms, in billions, on Xeon4.
Figure 10: Cache misses of parallel scoring algorithms, in billions, on Xeon4.
|
Table 1: Cache misses of scoring algorithms, in millions, on Xeon4.
Table 2: Run time of scoring algorithms, in hh:mm:ss, on Xeon4.
|
Figure 11 and Table 5 give the running times for our parallel scoring algorithms on our Xeon4 platform. In the figure, the time is in seconds while in the table, the time is given using the format hh : mm : ss. As in the table, PP_Strip is the fastest algorithm in practice, which is up to 40.0% faster than PP_Score and up to 38.4% faster than PP_Diagonal.
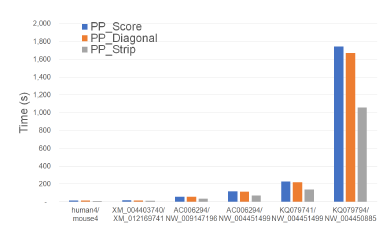 Figure 11: Run time of parallel scoring algorithms, in seconds, on Xeon4.
Figure 11: Run time of parallel scoring algorithms, in seconds, on Xeon4.
Table 6 gives the speedup of each of our parallel scoring algorithms relative to their sequential counterparts. As can be seen, the speedup of PP_Strip (i.e., Strip/PP_Strip) is between 3.92 and 3.98, which is quite close to the number of cores (4) on our Xeon4 platform. PP_Score achieves a speedup in the range
2.82 to 2.94 and the speedup for PP_Diagonal is in the range 3.12 to 3.21.
The excellent speedup exhibited by PP_Strip is due largely to our ability to greatly reduce cache misses for this algorithm.
Figures 12 and Tables 7 give the CPU and cache energy consumed, in joules, by our Xeon4 platform. On our datasets, PP_Strip required up to 41.2% less CPU and cache energy than PP_Score and up to 45.5% less than PP_Diagonal.
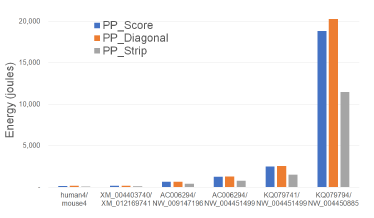 Figure 12: CPU and cache energy consumption of parallel scoring algorithms, in joules, on Xeon4.
Figure 12: CPU and cache energy consumption of parallel scoring algorithms, in joules, on Xeon4.
Compared to the sequential scoring algorithms, the multi-core algorithms use higher CPU power but less running time. Since the power increase is less than the decrease in running time, energy consumption is reduced.
4.2.3. Alignment Algorithms
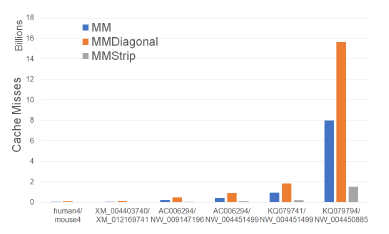 Figure 13: Cache misses for alignment algorithms, in billions, on Xeon4.
Figure 13: Cache misses for alignment algorithms, in billions, on Xeon4.
Figure 13 and Table 8 give the number of cache misses of our single-core alignment algorithms on our Xeon4 plat-
Table 3: CPU and cache energy consumption of scoring algorithms, in joules, on Xeon4.
| A | |A| | B | |B| | Score | Diagonal | Strip | Imp1 | Imp2 |
| human4 | 97,634 | mouse4 | 94,647 | 230.67 | 252.29 | 190.6 | 17.4% | 24.5% |
| XM_004403740 | 104,267 | XM_012169741 | 103,004 | 268.29 | 297.28 | 221.49 | 17.4% | 25.5% |
| AC006294 | 200,000 | NW_009147196 | 200,000 | 997.88 | 1100.6 | 829.36 | 16.9% | 24.6% |
| AC006294 | 200,000 | NW_004451499 | 398,273 | 2026.26 | 2178.97 | 1651.46 | 18.5% | 24.2% |
| KQ079741 | 392,981 | NW_004451499 | 398,273 | 3944.3 | 4300.59 | 3253.46 | 17.5% | 24.3% |
| KQ079794 | 1,083,068 | NW_004450885 | 1,098,196 | 30125.93 | 33472.81 | 24980.35 | 17.1% | 25.4% |
Table 4: Cache misses of parallel scoring algorithms, in millions, on Xeon4.
| A | |A| | B | |B| | PP_Score | PP_Diagonal | PP_Strip | Imp1 | Imp2 |
| human4 | 97,634 | mouse4 | 94,647 | 34 | 102 | 1 | 96.7% | 98.9% |
| XM_004403740 | 104,267 | XM_012169741 | 103,004 | 39 | 115 | 1 | 96.8% | 98.9% |
| AC006294 | 200,000 | NW_009147196 | 200,000 | 146 | 398 | 4 | 96.9% | 98.9% |
| AC006294 | 200,000 | NW_004451499 | 398,273 | 362 | 768 | 7 | 98.1% | 99.1% |
| KQ079741 | 392,981 | NW_004451499 | 398,273 | 628 | 1,373 | 14 | 97.7% | 99.0% |
| KQ079794 | 1,083,068 | NW_004450885 | 1,098,196 | 3,642 | 9,976 | 121 | 96.7% | 98.8% |
form. MMStrip has the fewest number of cache misses followed by MM and MMDiagonal (in this order). MMStrip reduces cache misses by up to 81.0% relative to MM and by up to 90.3% relative to MMDiagonal. Figure 14 and Table 9 give the running times of our single-core alignment algorithms on our Xeon4 platform. As can be seen, MMStrip is the fastest followed by MM and MMDiagonal (in this order). MMStrip reduces running time by up to 15.0% relative to MM, by up to 13.4% relative to MMDiagonal. As was the case with our scoring algorithms, the reduction in running time, while significant, isn’t as much as the reduction in cache misses.
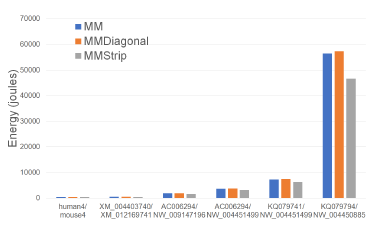 Figure 15: CPU and cache energy consumption of alignment algorithms, in joules, on Xeon4.
Figure 15: CPU and cache energy consumption of alignment algorithms, in joules, on Xeon4.
Figures 15 and Tables 10 give the CPU and cache energy consumption, in joules, by our single-core alignment algorithms. On our datasets, MMStrip reduced
up to 17.5% less CPU and cache energy than MM Figure 16: Cache misses for parallel alignment algoand up to 18.7% less than MMDiagonal. Once again, rithms, in billions, on Xeon4.
the energy reduction is comparable to the reduction in running time suggesting a close relationship between PP_MMStrip has the fewest number of cache running time and energy consumption for this applica- misses followed by PP_MM and PP_MMDiagonal tion. (in this order). PP_MMStrip reduces cache misses by Table 5: Run time of parallel scoring algorithms on Xeon4.
| A | |A| | B | |B| | PP_Score | PP_Diagonal | PP_Strip | Imp1 | Imp2 |
| human4 | 97,634 | mouse4 | 94,647 | 0:00:14 | 0:00:13 | 0:00:08 | 40.0% | 37.7% |
| XM_004403740 | 104,267 | XM_012169741 | 103,004 | 0:00:16 | 0:00:15 | 0:00:10 | 39.8% | 37.1% |
| AC006294 | 200,000 | NW_009147196 | 200,000 | 0:00:58 | 0:00:58 | 0:00:36 | 39.0% | 38.4% |
| AC006294 | 200,000 | NW_004451499 | 398,273 | 0:01:56 | 0:01:53 | 0:01:11 | 39.0% | 37.0% |
| KQ079741 | 392,981 | NW_004451499 | 398,273 | 0:03:49 | 0:03:39 | 0:02:19 | 39.3% | 36.6% |
| KQ079794 | 1,083,068 | NW_004450885 | 1,098,196 | 0:29:02 | 0:27:52 | 0:17:39 | 39.2% | 36.7% |
Table 6: Speedup of parallel scoring algorithms on Xeon4.
| A | |A| | B | |B| | Score/PP | Diagonal/PP | Strip/PP |
| human4 | 97,634 | mouse4 | 94,647 | 2.82 | 3.12 | 3.93 |
| XM_004403740 | 104,267 | XM_012169741 | 103,004 | 2.82 | 3.19 | 3.92 |
| AC006294 | 200,000 | NW_009147196 | 200,000 | 2.89 | 3.14 | 3.97 |
| AC006294 | 200,000 | NW_004451499 | 398,273 | 2.94 | 3.18 | 3.97 |
| KQ079741 | 392,981 | NW_004451499 | 398,273 | 2.89 | 3.21 | 3.98 |
| KQ079794 | 1,083,068 | NW_004450885 | 1,098,196 | 2.90 | 3.18 | 3.98 |
up to 95.5% relative to PP_MM and by up to 98.2% relative to PP_MMDiagonal.
Figure 17 and Table 12 give the running times for our parallel alignment algorithms on the Xeon4 platform. PP_MMStrip is faster than PP_MM by up to 37.4% and faster than PP_MMDiagonal by up to
40.3%.
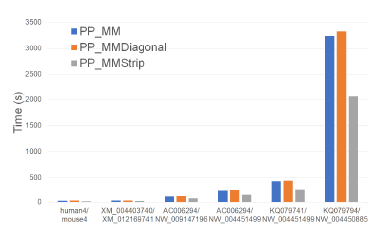 Figure 17: Run time of parallel alignment algorithms, in seconds, on Xeon4.
Figure 17: Run time of parallel alignment algorithms, in seconds, on Xeon4.
Table 13 gives the speedup of each parallel alignment algorithm relative to its single-core counterpart.
The speedup achieved by PP_MMStrip (relative to MMStrip) ranges from 3.56 to 3.94 while that for PP_MM is in the range 2.77 to 2.88 and that for PP_MMDiagonal is in the range 2.53 to 2.81.
Figures 18 and Tables 14 give the CPU and cache energy consumption, in joules, by our multi-core alignment algorithms. On our datasets, PP_MMStrip required up to 29.9% less CPU and cache energy than PP_MM and up to 42.1% less than PP_MMDiagonal. Once again, the energy reduction is comparable to the reduction in running time suggesting a close relationship between running time and energy consumption for this application.
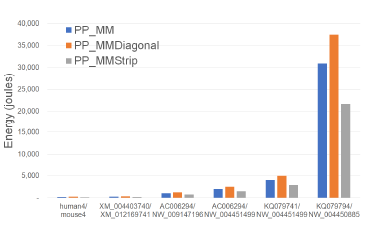 Figure 18: CPU and cache energy consumption of parallel alignment algorithms, in joules, on Xeon4.
Figure 18: CPU and cache energy consumption of parallel alignment algorithms, in joules, on Xeon4.
4.3. I7-x980 (Xeon6)
4.3.1. Scoring Algorithms
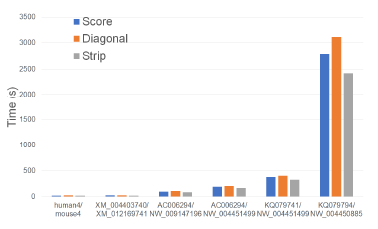 Figure 19: Run time of scoring algorithms, in seconds, on Xeon6
Figure 19: Run time of scoring algorithms, in seconds, on Xeon6
Figure 19 and Table 15 give the running times of our single-core scoring algorithms on our Xeon6 platform. As can be seen, Strip is the fastest followed by Score and Diagonal (in this order). Strip reduces running time by up to 14.3% relative to Score and by up to 22.4% relative to Diagonal.
|
Table 7: CPU and cache energy consumption of parallel scoring algorithms on Xeon4.
Table 8: Cache misses for alignment algorithms, in millions, on Xeon4.
|
4.3.2. Parallel Scoring Algorithms
Figure 20 and Table 16 give the running times for our parallel scoring algorithms on our Xeon6 platform. As with Xeon4, PP_Strip is faster than PP_Score and PP_Diagonal and reduces the running time by up to 42.5% and 55.6%, respectively.
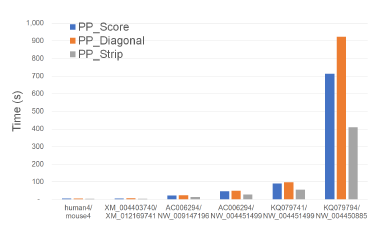 Figure 20: Run time of parallel scoring algorithms, in seconds, on Xeon6.
Figure 20: Run time of parallel scoring algorithms, in seconds, on Xeon6.
Table 18 gives the speedup of each of our parallel algorithms relative to their single-core counterparts. PP_Strip achieves a speedup of up to 5.89, which is very close to the number of cores. The maximum speedup achieved by PP_Score and PP_Diagonal was 4.09 and 4.25, respectively.
4.3.3. Alignment Algorithms
Figure 21 and Table 17 give the running times of our parallel scoring algorithms on the Xeon6 platform. As can be seen, MMStrip is the fastest followed by MM and MMDiagonal (in this order). MMStrip reduces running time by up to 12.6% relative to MM and by up to 14.2% relative to MMDiagonal.
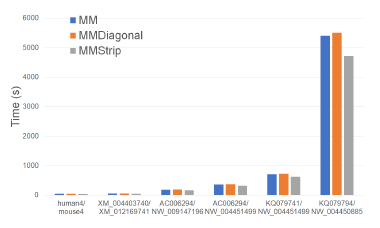 Figure 21: Run time of alignment algorithms, in seconds, on Xeon6.
Figure 21: Run time of alignment algorithms, in seconds, on Xeon6.
4.3.4. Parallel Alignment Algorithms
Figure 22 and Table 20 give the running times of our parallel alignment algorithms on the Xeon6.
PP_MMStrip is faster than PP_MM and PP_MMDiagonal and reduces the running time by up to 39.9% and 44.8%, respectively.
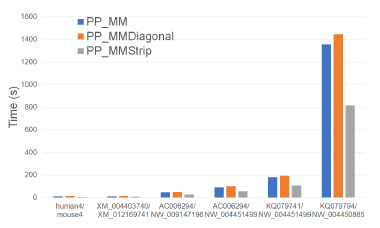 Figure 22: Run time of parallel alignment algorithms, in seconds, on Xeon6.
Figure 22: Run time of parallel alignment algorithms, in seconds, on Xeon6.
|
Table 9: Run time of alignment algorithms on Xeon4.
Table 10: CPU and cache energy consumption of alignment algorithms on Xeon4.
|
Table 21 gives the speedup of each of our parallel algorithms relative to their single-core counterparts. PP_MMStrip achieves a speedup of up to 5.78, which is very close to the number of cores.
The maximum speedup achieved by PP_MM and PP_MMDiagonal was 3.98 and 3.80, respectively.
4.4. Xeon E5-2695 (Xeon24)
4.4.1. Scoring Algorithms
Figure 23 and Table 19 give the running times of our single-core scoring algorithms on our Xeon24 platform. As was the case on our other test platforms, Strip is the fastest followed by Score and Diagonal (in this order). Strip reduces running time by up to 19.7% relative to Score and by up to 35.1% relative to Diagonal.
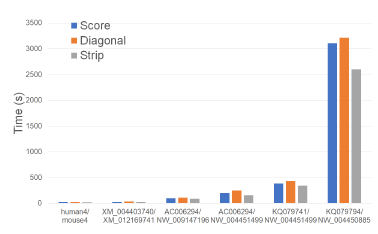 Figure 23: Run time of scoring algorithms, in seconds, on Xeon24.
Figure 23: Run time of scoring algorithms, in seconds, on Xeon24.
4.4.2. Parallel Scoring
Algorithms Figure 24 and Table 22 give the running times for our parallel scoring algorithms on our Xeon24 platform. PP_Strip is faster than PP_Score and PP_Diagonal and reduces the running time by up to 61.4% and 76.2%, respectively.
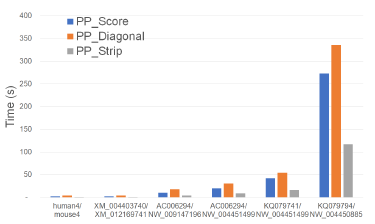 Figure 24: Run time of parallel scoring algorithms, in seconds, on Xeon24.
Figure 24: Run time of parallel scoring algorithms, in seconds, on Xeon24.
Table 23 gives the achieved speedup. PP_Strip scales quite well and results in a speedup of up to 22.22.
The maximum speedups provided by PP_Score and PP_Diagonal are 11.36 and 9.56, respectively.
4.4.3. Alignment Algorithms
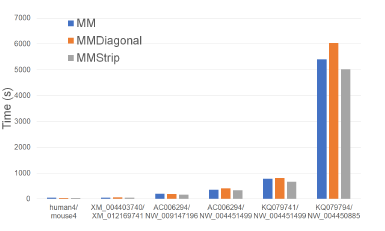 Figure 25: Run time of alignment algorithms, in seconds, on Xeon24.
Figure 25: Run time of alignment algorithms, in seconds, on Xeon24.
Figure 25 and Table 24 give the running times of our single-core alignment algorithms on our Xeon24 plat-
|
Table 11: Cache misses for parallel alignment algorithms, in millions, on Xeon4.
Table 12: Run time of parallel alignment algorithms on Xeon4.
|
form. MMStrip is the fastest followed by MM and MMDiagonal (in this order). MMStrip reduces running time by up to 17.1% relative to MM and by up to 16.8% relative to MMDiagonal.
4.4.4. Parallel Alignment Algorithms
Figure 26 and Table 25 give the running times of our parallel alignment algorithms on Xeon24. As can be seen, PP_MMStrip is faster than PP_MM and PP_MMDiagonal. It reduces the running time by up to 47.3% and 84.6%, respectively.
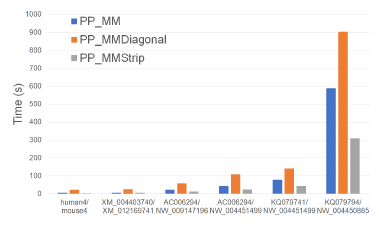 Figure 26: Run time of parallel alignment algorithms, in seconds, on Xeon24.
Figure 26: Run time of parallel alignment algorithms, in seconds, on Xeon24.
Table 26 gives the speedup of our parallel algorithms. PP_MMStrip achieves a speedup of up to 16.2 while PP_MM and PP_MMDiagonal have maximum speedups of 9.79 and 6.58.
5. Discussion
By accounting for the presence of caches in modern computers, we are able to arrive at sequence alignment algorithms that are considerably faster than those that do not take advantage of computer caches. Our benchmarking demonstrates the value of optimizing cache usage. Our cache-efficient algorithms Strip and MMStrip were the best-performing single-core algorithms and their parallel counterparts were the bestperforming parallel algorithms. Strip reduced running time by as much as 19.7% relative to the classical scoring algorithm Score due to Smith and Waterman and
MM_Strip reduced running time by as much as 17.1% relative to the alignment algorithm of Myers and Miller. Neither the algorithm of Smith and Waterman nor that of Myers and Miller optimize cache utilization. The parallel versions of Strip and MM_Strip were up to 61.4% and 47.3% faster than the parallel versions of the Smith and Waterman and the Myers and Miller algorithms, respectively.
6. Limitations
Our cache miss analyses assume a simple cache model in which there is a single LRU cache. In practice, computers have multiple levels of cache and employ sophisticated and proprietary cache replacement strategies. Despite the use of a simplified cache model for analysis, the developed cache-efficient algorithms perform very well in practice.
7. Conclusion
The main contributions of this papers are
- cache efficient single-core and multi-core algorithms to determine the score of the best alignment;
- cache efficient single-core and multi-core algorithms to determine the best alignment.
The effectiveness of our cache-efficient algorithms has been demonstrated experimentally using three computational platforms. Future work includes developing the cache-efficient algorithms for other problems in computational biology.
Conflict of Interest The authors declare no conflict of interest.
Table 13: Speedup of parallel alignment algorithms on Xeon4.
| A | |A| | B | |B| | MM/PP | MMDiagonal/PP | MMStrip/PP |
| human4 | 97,634 | mouse4 | 94,647 | 2.77 | 2.53 | 3.70 |
| XM_004403740 | 104,267 | XM_012169741 | 103,004 | 2.78 | 2.56 | 3.56 |
| AC006294 | 200,000 | NW_009147196 | 200,000 | 2.80 | 2.70 | 3.81 |
| AC006294 | 200,000 | NW_004451499 | 398,273 | 2.84 | 2.69 | 3.89 |
| KQ079741 | 392,981 | NW_004451499 | 398,273 | 2.84 | 2.78 | 3.94 |
| KQ079794 | 1,083,068 | NW_004450885 | 1,098,196 | 2.88 | 2.81 | 3.93 |
Table 14: CPU and cache energy consumption of parallel alignment algorithms on Xeon4.
| A | |A| | B | |B| | PP_MM | PP_MMDiagonal | PP_MMStrip | Imp1 | Imp2 |
| human4 | 97634 | mouse4 | 94647 | 236.98 | 305.14 | 181.6 | 23.4% | 40.5% |
| XM_004403740 | 104267 | XM_012169741 | 103004 | 272.26 | 352.56 | 216.83 | 20.4% | 38.5% |
| AC006294 | 200000 | NW_009147196 | 200000 | 1044.86 | 1279.77 | 747.86 | 28.4% | 41.6% |
| AC006294 | 200000 | NW_004451499 | 398273 | 2039.55 | 2540.52 | 1483.8 | 27.2% | 41.6% |
| KQ079741 | 392981 | NW_004451499 | 398273 | 4020.48 | 4979.17 | 2905.32 | 27.7% | 41.7% |
| KQ079794 | 1083068 | NW_004450885 | 1098196 | 30964.02 | 37461.35 | 21703.65 | 29.9% | 42.1% |
Acknowledgment
This work was supported, in part, by the National Science Foundation under award NSF 1447711.
- Henikoff and J. G. Henikoff, “Amino acid substitution matrices from protein blocks,” Proc Natl Acad Sci U S A, vol. 89, pp. 10 915–10 919, 1992.
- B. Needleman and C. D. Wunsch, “A general method applicable to the search for similarities in the amino acid sequence of two proteins,” Journal of Molecular Biology, vol. 48, pp. 443–453, 1970.
- F. Smith and M. S. Waterman, “Identification of common molecular subsequences,” Journal of Molecular Biology, vol. 147, pp. 195–197, 1981.
- Gotoh, “An improved algorithm for matching biological sequences,” Journal of Molecular Biology, vol. 162, pp. 705–708, 1982.
- S. Hirschberg, “A linear space algorithm for computing longest common subsequences,” Communications of the ACM, vol. 18, pp. 341–343, 1975.
- Myers and W. Miller, “Optimal alignments in linear space,” Computer Applications in the Biosciences(CABIOS), vol. 4, pp. 11–17, 1988.
- Huang, R. Hardison, and W. Miller, “A space-efficient algorithm for local similarities,” Comput Appl Biosci, vol. 6, p. 373âAS381, 1990.
- Altschul, W. Gish, W. Miller, E. Myers, and D. Lipman, “Basic local alignment search tool,” Journal of Molecular Biology, vol. 215, pp. 403–410, 1990.
- Pearson and D. Lipman, “Improved tools for biological sequence comparison,” Proceedings of the National Academy of Sciences USA, vol. 85, pp. 2444–2448, 1988.
- Chao, J. Zhang, J. Ostell, and W. Miller, “A local alignment tool for very long dna sequences,” Comput Appl Biosci, vol. 11, pp. 147–153, 1995.
- Driga, P. Lu, J. Schaeffer, D. Szafron, K. Charter, and I. Parsons, “Fastlsa: a fast, linear-space, parallel and sequential algorithm for sequence alignment,” Algorithmica, vol. 45, p. 337âAS375, 2006.
- Li, S. Ranka, and S. Sahni, “Pairwise sequence alignment for very long sequences on gpus,” IEEE 2nd International Conference on Computational Advances in Bio and Medical Sciences (ICCABS), 2012.
- O. Sandes and A. C. M. A. Melo, “Smith-waterman alignment of huge sequences with gpu in linear space,” IEEE International Symposium on Parallel and Distributed Processing (IPDPS), pp. 1199–1211, 2011.
- Aluru and N. Jammula, “A review of hardware acceleration for computational genomics,” IEEE Design and Test, vol. 31, pp. 19–30, 2014.
- Rajko and S. Aluru, “Space and time optimal parallel sequence alignments,” IEEETPDS:IEEE Transactions on Parallel and Distributed Systems, vol. 15, 2004.
- Khajeh-Saeed, S. Poole, and J. B. Perot, “Acceleration of the smithâASwaterman algorithm using single and multiple graphics processors,” Journal of Computational Physics, p. 4247âAS4258, 2010.
- Kurtz, A. Phillippy, A. L. Delcher, M. Smoot, M. Shumway, C. Antonescu, and S. L. Salzberg, “Versatile and open software for comparing large genomes,” Genome Biol, vol. 5, 2004.
- Almeida and N. Roma, “A parallel programming framework for multi-core dna sequence alignment,” Complex, Intelligent and Software Intensive Systems (CISIS), 2010 International Conference on, pp. 907 – 912, 2010.
- Hamidouche, F. M. Mendonca, J. Falcou, A. C. M. A. Melo, and D. Etiemble, “Parallel smith-waterman comparison on multicore and manycore computing platforms with bsp++,” International Journal of Parallel Programming, vol. 41, pp. 1110–136, 2013.
- Zhao and S. Sahni, “Cache and energy efficient alignment of very long sequences,” 2015 IEEE 5th international conference on Computational Advances in Bio and Medical Sciences (ICCABS), 2015.
- “Perf tool,” https://perf.wiki.kernel.org/index.php/ Main_Page.
- “Globin gene server,” http://globin.cse.psu.edu/globin/ html/pip/examples.html.
- “Ncbi database,” http://www.ncbi.nlm.nih.gov/gquery.

