Improving Patient Outcomes Through Untethered Patient-Centered Health Records
Volume 3, Issue 2, Page No 164-173, 2018
Author’s Name: Mohammed Abdulkareem Alyami1,a), Majed Almotairi1, Alberto R. Yataco2, Yeong-Tae Song1
View Affiliations
1Dept. of Computer & Information Sciences, Towson University, 21252, USA
2Medical Director Get Well Immediate Care Towson, 21204, USA
a)Author to whom correspondence should be addressed. E-mail: aljakman2006@hotmail.com
Adv. Sci. Technol. Eng. Syst. J. 3(2), 164-173 (2018); ![]() DOI: 10.25046/aj030219
DOI: 10.25046/aj030219
Keywords: Personal Health Records (PHRs), Personal Health Records Systems (PHRS), Dublin Core Metadata, Cloud storage, Standard Medical Codes
Export Citations
Patient generated data, or personal clinical data, is considered an important aspect in improving patient outcomes. However, personal clinical data is difficult to collect and manage due to its distributed nature. For example, they can be located in multiple places such as doctors’ offices, radiology centers, hospitals, or some clinics. Another factor that can make personal clinical data difficult to manage is that it can be heterogeneous data types such as text, images, charts, or paper-based documents. In case of emergencies, this situation makes personal clinical data retrieval very difficult. In addition, since the amount and types of personal clinical data continue to grow, finding relevant clinical data when needed is getting more difficult if no action is taken. In response to such scenarios, we propose an untethered patient health record system that manages personal health data by utilizing meta-data that enables easy retrieval of clinical data. We incorporate cloud-based storage for easy access and sharing with caregivers to implement continuity of care and evidence-based treatment. In emergency cases, we make critical medical information such as current medications and allergies available to relevant caregivers with valid license numbers only. Clinical data needs to be stored or made accessible from one place for easy sharing and retrieval. Well-managed personal cloud space could outlive the lifetime of personal health records system (PHRS) since the discontinuity of the service does not affect the data stored in the cloud space. In our approach, we separate the clinical data from applications in order to make the data independent from the application. Also, the users can have alternative applications for their clinical data. Such independence motivates users to use PHRS with flexibility.
Received: 16 November 2017, Accepted: 11 March 2018, Published Online: 27 March 2018
1. Introduction
For most people, healthcare is considered important as there has been significant increase in chronic diseases such as heart disease, cancer, diabetes and asthma. This requires continuous treatment, reduces quality of life, and increases overall medical expenses (The Growing Crisis of Chronic Disease in the United States) [1]. According to the Centers for Disease Control and Prevention (CDC), in the U.S. about 610,000 people die of heart disease every year. In addition, 26 million people suffer from Type I or Type II Diabetes, around 14 million have severe chronic respiratory problems such as Chronic Obstructive Pulmonary Disease (COPD), and 68 million have been diagnosed with hypertension [2]. However, many of these diseases can be prevented and managed through early detection, physical activities, a balanced diet and treatment therapy. Adopting PHRS could help improve patient outcomes. Recently, there has been more focus on preventive care and proactive measures such as monitoring and controlling patients’ symptoms. Nowadays, there are many mobile health applications and sensors such as blood pressure sensors, electrocardiogram sensors, blood glucose measuring devices, and others that are used for monitoring and controlling personal health. These apps and sensors produce personal health data that can be used for treatment purposes. If managed and handled properly, it can be considered patient-generated data. There are other types of personal health data that are available from various sources such as hospitals, doctor’s offices, clinics, radiology centers or any other caregivers. Aforementioned health documents are deemed as a personal health record (PHR). According to American Health Information Management Association (AHIMA) [3], PHR can be defined as an electronic, lifelong resource of health information needed by individuals to make health decisions. However, it is not easy to collect all the relevant personal health data because of the fact that they are in different data types, available from different sources, and stored in different media and devices. To overcome such difficulties, it is desirable to have personal health data in one place where users have full control over their own clinical data. In order to be useful, the clinical data should be sharable when needed for diagnosis and treatment. Without proper clinical information (medical history, allergies, current medications, and adverse reactions) medical mistakes could occur when making medical decisions due to insufficient information. Even if a patient has a complete medical history and all the necessary clinical data, if it is not shared properly among caregivers at the time of need, discontinuity in care may occur. In order to meet the needs of such scenario, PHRS should have the following properties: robust and private storage, easy retrieval and maintenance, secure, sharable, and able to handle emergency situations.
There are two types of PHRS: untethered and tethered. Untethered PHR is an independent PHRS where patients have full control over their own personal health records. They can collect, manage, and share their health records. On the other hand, tethered PHRS are linked to a specific healthcare providers’ EHR system, where the users typically gain easy access to their own records through secure portals and see their own clinical information such as test results, immunization records, family history, and other relevant information. They can also utilize secure messaging with their collaborating clinicians. The participating patients need to share the cost and the information with their care provider. However, these tethered records may not be complete since the information sources are from one provider only. Despite all the benefits PHRS provide, the adoption rate of PHR by the general public still remains low in the U.S. [4-6]. In our previous work [7], we identified six barriers (usability, ownership, interoperability, privacy and security, portability and motivation) that cause the slow adoption of PHRS. One of the main concerns for not having PHRS is the ownership issue. For example, one incident happened on January 1, 2012. Google Health™ System, one of the biggest PHRS providers, stopped its service and asked their registered customers to move their records to their computers or other PHRS vendors, which exacerbated ownership concerns by the public.
As an attempt to overcome some of the barriers, we propose an untethered PHRS that utilizes personal cloud storage, offers simplicity in organizing various kinds of clinical data by utilizing Dublin Core (DC) metadata, and provides easy access to emergency clinical data to paramedics or clinicians in case of emergency. DC metadata has been successfully applied in many areas, but since it is not specifically designed for clinical data, there are some limitations in its expressive power in the healthcare domain. In this research, we simplified the categorization of clinical data by human body part for easy retrieval of clinical data using DC, so users can manage their own clinical data without in-depth knowledge about clinical information. As a proof of our concept, we developed a system called My Clinical Record System (MCRS) to help users store, organize, retrieve, and share their clinical data with caregivers when needed, including in emergency situations. In an emergency situation, clinicians (e.g. physicians, paramedics, nurses, and others) can access patient’s data using their license numbers and the patient’s name and date of birth. Emergency information consists of current medication lists, known allergies, and side effects. By having complete medical history, MCRS users may be able to reduce medical errors and improve patients’ outcomes. It also ensures continuity of care by sharing personal clinical data among healthcare providers when needed. The remainder of this paper is organized as follows: Section 2 discusses related work and in section 3 we focus on the clinical data’s type and format. In Section 4, we discuss how to solve the data organization and retrieval issues using DC metadata to facilitate better and more accurate data retrieval. Section 5 discuses cloud-based storage. In section 6, we discuss the current situation in the healthcare industry (AS-IS). Section 7 uses a scenario as an example. Section 8 discusses the solution domain. In section 9, we introduce MCRS as a proof of concept and finally we conclude our study and discuss our future work in section 10.
2. Literature Review
In [8], Fearon defined Meta-data as structured information that describes, explains, locates, or otherwise makes it easier to retrieve, use, or manage an information resource. However, metadata standards have not been employed by many repositories and most of the meta-data was generally descriptive, rather than administrative or for preservation [9]. In [10], Greenberg investigated the ability of resource authors to create acceptable metadata in an organizational web site at the National Institute of Environmental Health Sciences in the U.S. The findings of their study indicate that resource authors can create better quality metadata when working with Dublin Core. In some circumstances, they may be able to create quality metadata better than a metadata professional. In [11], Talha developed their own metadata tool called Metadata Management System (MMS) to facilitate the creation, maintenance and storage of metadata. MMS supports two well-known metadata models, Dublin Core and SCORM 1.2 (IEEE Learning Object Metadata). The authors implemented their metadata tools in the Malaysia Grid for Learning (MyGfL). The results of the study indicate that MMS can substantially improve the discovery, retrieval management and control of web resources and learning objects in their MyGfL portal. In the healthcare domain, meta-data has been utilized as a method for confidentiality tags that indicate data sensitivity levels. This enables patients to give consent to the exchange of some parts of their health records (e.g. the medical diagnosis), while withholding consent for the exchange of other areas (e.g. a mental health counseling session) [12]. This work can help in increasing patients’ privacy and allow them with some freedom in exchanging their health records. However, their work focused on EHR, but did not incorporate PHRS, which limited the scope of their study. Other researchers have adopted the ontology approach to quickly search and access relevant and meaningful information among large numbers of CDA documents within healthcare providers’ systems (Electronic Health System), which in turn enables semantic interoperability[5, 13]. However, these studies were limited to one health data type (CDA), whereas health records include other data types such as images (e.g. x-rays, scanned documents, ultrasounds, and others), and observed symptoms noted by patients and clinical sensors’ data. In [14] Patel developed a system called TrialX, on top of PHR, where patients not only can search by keywords, as in ClinicalTrials.gov, but also by demographics (e.g. age, gender, city and study site). This system enables patients to match their health condition to clinical trials. This system can accelerate and improve the search results among health records. However, the authors did not take into account how to retrieve relevant clinical data since the amount and types of personal clinical data continue to grow, which makes it difficult to find such data. In [15], Appelboom reviewed the literature on smart wearable body sensors and found that these sensors are accurate and have clinical utility, but still are underutilized in the healthcare industry. These devices can be used to monitor physiological, cardiovascular and many other factors of health variables and transmit data either to a personal device or to an online storage site. The smart wearable body sensors are placed on different parts of the user’s body based on the purpose of the sensor device. For instance, the physical therapy sensor is placed on the ankle; the cardiopulmonary sensor can be placed on the wrists, fingers, arms or thighs.
In [16], Zhang developed an application to apply meta-data efficiently on clinical trial data. The authors chose Microsoft Excel due to the wealth of built-in features (e.g. spell checking, sorting, filtering, finding, replacing, importing and exporting data capabilities), which contribute to the ease of use, power, and flexibility of the overall meta-data application. They focused on the analysis process in a drug development environment such as adverse clinical events (ACE), Electrocardiogram (ECG), laboratories (LAB), and vital signs (VITAL), where the raw data is stored in the clinical trial database and then the data can be manipulated.
Another study in [17], Teitz developed a website called HealthCyberMap with the goal of mapping Internet health information resources in novel ways for enhanced retrieval and navigation. They used Protégé-2000 to model and populate a qualified DC RDF meta-data base. They also extended the DC elements by adding quality and location elements. Also, the W3C RDFPic project extends the DC schema by adding its own elements such as camera, film, lens and film development date for describing and retrieving digitized photos. In [18], Ekblaw built a system (RedRec) to enable patients to access their medical health records across health providers (e.g. pediatrician, university physician, dentist, employer health plan provider, specialists, and others). Their system applies novel, blockchain smart contracts to create a decentralized content-management system for healthcare data across providers. RedRec governs medical records access while providing the patient with the ability to share, review, and post new records via flexible interface. The raw medical record content is kept securely in providers’ existing data storage. However, when the patient wants to retrieve data from their provider’s database, their Database Gatekeeper checks authentication. If it is approved, the Gatekeeper retrieves the relevant data for the requester and allows a sync with the local database.
In order to solve interoperability problems in exchanging clinical data, in [19], Dogac proposed archetypes to overcome the interoperability problems. They provided guidelines on how ebXML Registries can be used as an efficient medium for annotating, storing, discovering and retrieving archetypes. They also used ebXMLWeb services to retrieve data from clinical information systems. An archetype is defined as “a reusable, formal expression of a distinct, domain-level concept such as “blood pressure”, “physical ex-amination”, or “laboratory results”, expressed in the form of constraints on data whose instances conform to some reference model”[19]. However, these studies are not comprehensive. Their limitations come from: focus only on one health data type, do not take into account PHR, lack of retrieving relevant clinical data, and overlook emergency access to medical records. Our proposed approach overcomes these limitations. Our study is comprehensive, which covers many different clinical and nonclinical documents such as images (e.g. x-rays, scanned documents, ultrasounds, and others), text (e.g. CDA, CCR, CCD, and others), and observed symptoms noted by patients and their clinical sensors’ data. This can organize these various data types in a way that can help in storing and retrieving such data in an efficient way.
3. Clinical Data
In this section, we describe the types, formats, and sources of clinical data.
3.1. Measurements data from portable medical devices, sensors, or mobile application
One way to collect measurement data is through explicit clinical sensors. A clinical sensor is a device that responds to a physical stimulus and transmits a resulting impulse for interpretation or recording. Some sensors are designed to work outside the human body, while others can be implanted within the human body [20]. In this research, we are referring to clinical sensors for homecare settings, such as blood oxygen monitors, thermometers for body temperature, heart rate sensors, blood pressure sensors, and others. In addition to these textual data types, there can be non-textual data generated from sensors such as electrocardiogram measurement devices. The clinical sensors play a major role in healthcare, including early detection of diseases, diagnosis, disease monitoring and treatment monitoring.
Another method to collect measurement data is through mobile apps. For instance, most smartphones (e.g. Android or IOS) offer health and fitness apps that help users monitor their daily activities and health (e.g. track diet and nutrition calories, track vital signs, track fitness progress, share health data with their doctor electronically, and others). The data collected from these applications can be sent as a message or an email attachment to whom the users want to share it with. For interoperability, the collected data needs to be in standard format, such as HL7 CDA or in standard code such as SNOMED-CT.
3.2. Observed Symptoms
Patients sometimes experience particular symptoms (e.g. chest pain, nausea, vomiting, shortness of breath and others). If the patient notices such symptoms, they should be recorded and shared with their physician for proper treatment. If these symptoms are not shared with their physician, due to incomplete information, misdiagnosis could occur. When patients are recording, the observed symptoms should be described in standardized code such as SNOMED-CT. This will allow semantic interoperability, since the same symptoms can be described in multiple ways. Without codified descriptions, there can be discrepancies about the perceptions regarding symptoms between patients, nurses, or physicians [21].
3.3. Images
Most of the medical imaging machines produce standard image format called Digital Imaging and Communications in Medicine (DICOM). DICOM is defined as the international standard for medical images and related information (ISO 12052). There are two types of clinical data images: images that are based on DICOM standard (e.g. x-rays, Computed Tomography (CT), Magnetic Resonance (MR), and ultrasound devices) and scanned documents. The DICOM –format combines images and meta-data that describes the medical imaging procedure. Accessing data in DICOM files becomes as easy as working with TIFF or JPEG images [22]. On the other hand, the scanned documents (e.g. PDF/ JPEG) are difficult to retrieve because the content is not searchable. For example, some physicians write notes on clinical forms while diagnosing their patients and then type them on the computer. They also sometimes scan the notes and upload them to the patient records. Either way is time consuming, difficult to retrieve in a timely manner, and consumes relatively large storage space. In addition, the patient may have more than one doctor or may have been treated by many healthcare providers, which in turn fragments his/her records. So when patients obtain their records, they mostly receive them either printed out or sent as an email attachment. This makes it difficult to retrieve scanned documents because its content cannot be retrieved by computers. To alleviate such issues, we have utilized meta-data to describe such medical documents so computerized retrieval and systematic organization are possible.
3.4. Clinical Document
Clinical documentation (CD) is a computerized record describing a medical treatment, medical trial or clinical test, which can be exchanged among healthcare providers [23]. EHR data may be collected from healthcare providers. There are four types of clinical document formats: Continuity of Care Record (CCR), Clinical Document Architecture (CDA), Continuity of Care Document (CCD), and Consolidated CDA (C-CDA). All of which allow healthcare providers to exchange clinical information summary about a patient. However, CCR was excluded from the 2014 edition of EHR Certification, which is a standard certification criteria for EHRs that was established by The Centers for Medicare & Medicaid Services (CMS) and the Office of the National Coordinator for Health Information Technology (ONC), as a valid way to send summary of care documents. Hence, the content from a CCR was merged into a CDA format and called Continuity of Care Document (CCD). C-CDA includes nine different types of commonly used CDA documents such as CCD, consultation notes, discharge summary, imaging integration, DICOM diagnostic imaging reports, history and physical, operative note, progress note, procedure note, and unstructured documents. Each C-CDA Document Template was designed to satisfy a specific information exchange situation.
In the following subsection, we will discuss the difference between these formats (CCR, CDA, and CCR):
- CCR documents provide a snapshot of treatment and basic patient information such as diagnosis and reason for referral between healthcare providers. It also uses only specified XML code and does not support narrative text
(free-text), which hinders physicians from writing notes if needed, and it is not electronically acceptable by all systems.
- CDA documents are a flexible standard which can be read by the human eye or processed by machine due to the use of XML language. It is based on the HL7 Reference Information Model (RIM), and uses HL7 V3 data types. CDA can be transported using different methods such as HL7 V2, HL7 V3, DICOM, MIME-encoded attachments, HTTP, or FTP. CDA documents can include text, images and even multimedia.
- CCD documents are not a complete medical history of the patient but it includes only the information critical to effectively provide continuity of care. Its primary purpose is exchanging patient information between different healthcare providers. It is based on XML standard and can be displayed on a web browser using style sheet. It also allows narrative text which is an advantage over other standards like CCR [24].
When using untethered PHRS, patients are responsible for collecting clinical data from their healthcare providers or from their own patient-generated measurement data and keeping it in their own personal cloud space. For example, CDA, CCR and CCD can be obtained from healthcare providers, X-rays can be obtained from the radiology department, and lab test results can be obtained from the test lab or doctor’s office. Patients can share their health records with their clinicians by either electronically transmitting or granting access to their storage through the PHRS. If electronic sharing is not allowed, the patient may download the file and make hardcopies or store them in a USB, CD, or other mediums for sharing [25].
3.5. Meta-data
There are two different methods of storing meta-data. In the first method, meta-data can be embedded in the data (e.g. in the header of a digital file). The advantages of this option are ensuring that the meta-data will not be lost, eliminating the need for linking data and meta-data, and updating the data and meta-data together. In the second method, meta-data can be stored separately. The advantage of this option is that it can simplify the management of meta-data and can expedite the retrieval of the data [8]. In our approach, we employed the latter method to accelerate the retrieval of clinical data and to enhance expressive power. However, in this method, there can be inconsistencies between meta-data and clinical data. This can occur when transitioning to a new platform, integration between different systems or sharing data across multiple systems [12]. There are many meta-data formats that have been accepted internationally including: Dublin Core (DC), Federal Geographic Data Committee (FGDC), Encoded Archival Description (EAD), and Government Information Locator Service (GILS) [26]. Some of the metadata standards/schemas are generic, while others are domain-specific. For example , SCORM 1.2 (IEEE LOM) is used for educational approaches and rights management; Friend of a Friend (FOAF) is used for people and organizations; Simple Knowledge Organization (SKOS) is used for concept collections; Asset Description Metadata Schema (ADMS) is used for describing semantic interoperability assets; and Dublin Core is used for published material (text, images).
Generic metadata formats, such as Dublin Core, tend to be easy to use and widely adopted. In this study, we adopted DC metadata because it was developed for author-generated metadata, supports resource sharing and interoperability among information systems, has the broadest level of commonality of elements, extensibility, international acceptability and the flexibility it provides for extensions to the basic elements.
DC metadata solves one of the major issues in using meta-data among healthcare systems, which lacks interoperability [7]. Therefore, among these different metadata formats, DC metadata is the most appropriate format that aligns with our approach.
Meta-data benefits personal health record management in many ways. These benefits include the following:
- Consistency in definitions: properly defined tags provide structured information about the clinical data stored.
- Clarification of the relationships: meta-data can be used to clarify the relationships among the clinical items by defining categories and associated relationships within the category. We have defined each tag in DC for clinical purposes. When the data is uploaded or modified, the corresponding meta-data is required to update as well.
3.6. Meta-data Management
Meta-data management ensures that the data is associated with the datasets and utilized efficiently throughout and across organizations. Data governance is needed for successful meta-data implementation so it can provide trustworthy, timely and relevant information to decision makers, as well as personal users. For successful implementation, data governance must be aligned with the intended purposes of the users or organizations. This means that a data governance program starts by specifying its strategy, goals and the scope of its success. An organization needs to define the three data governance pillars including policies, people (and people skills), and processes. Once the above steps are in place, the organization can determine the best tools and technology to implement its data governance initiative [27].
3.7. Dublin Core Meta-data for Clinical Use
The DC Meta-Data Initiative (DCMI), is an open organization supporting innovation in meta-data design and best practices across the meta-data ecology. The DC Meta-data consists of 15 optional elements including: title creator, subject, description, publisher, contributor, date, type, format, identifier, source, language, relation, coverage and rights.
In this study, we defined the usage of DC Meta-data elements for clinical purposes to describe and retrieve clinical data efficiently as shown in Table 1. Some of the meta-data elements – title subject, description, type, data and resource – are required for the integrity of data. These elements must be present for every clinical data item. The optional fields can be skipped, but if it has been filled, the metadata quality will be increased.
Table 1: Meta-data schema for PHR
| Entity | Description |
| DC. Title | The title of the document |
| DC. Creator | The author of the document |
| DC. Subject | Subject of the document |
| DC. Description | Description of the document |
| DC. Relation |
– One of the body parts (Thorax, Abdomen, Heart, extremities, Integumentary, Head, Urinary or Reproductive) – This element is linked to the subject element |
| DC. Date | Date of meta data creation |
| DC. Type | Type will be used for lab tests (blood work, urinalysis, fecal sample, nasopharyngeal sample, oropharyngeal sample and others) or images (x-ray, cat scan/ CT, Ultrasound, Magnetic resonance/MR, Scanned Document, Electrocardiogram, EKG/ or ECG, CDA, C-CCD and others) |
| DC. Format | PDFs, Text, JPEGs , TIFFs, HL7 CDA, and others |
| DC. Identifier | Optional Document ID |
| DC. Language | English and other languages |
| DC. Coverage | Geographical and time-related information |
| DC. Rights | Copyright and access rights (secured or unsecured) |
| DC. Source | Data source |
4. Healthcare Processes
4.1. AS-IS Healthcare Processes
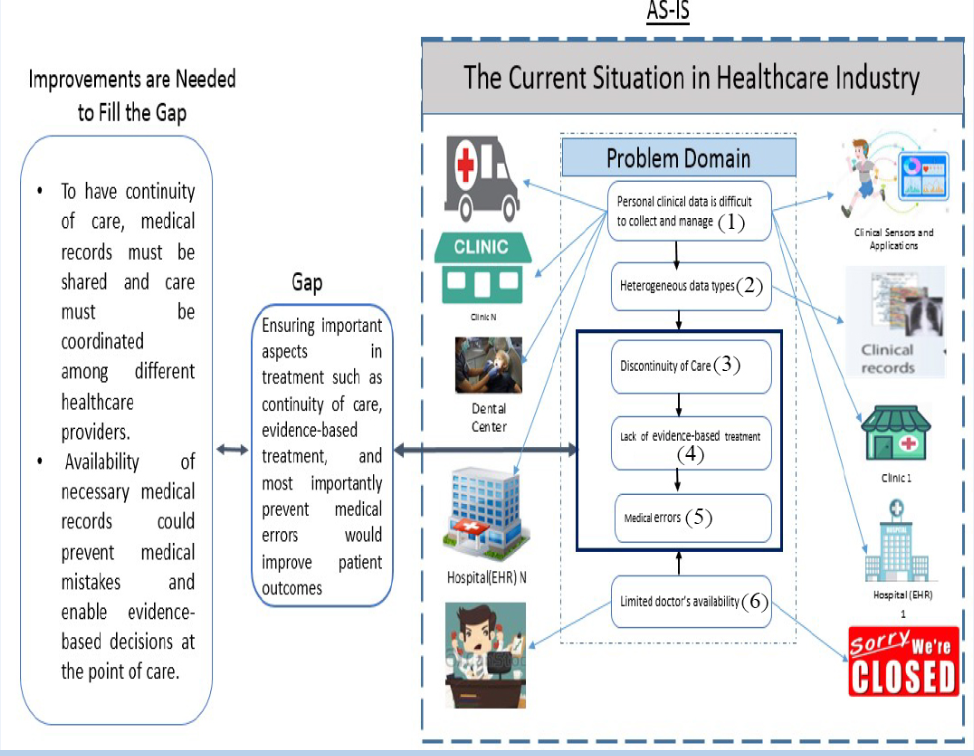
Some of the issues that the current healthcare industry is having include discontinuity of care and unacceptably high rates of medical mistakes due to unavailable patient medical records at the time of need. Some of the factors that cause these issues are listed below and illustrated in the Figure 1. (the numbers listed below correspond to the numbers in the Figure 1)
- Personal clinical data is difficult to collect and manage because they are located over multiple places such as doctor’s offices, radiology centers, hospitals, or some clinics (1).
- Heterogeneous data types such as text, images, charts, or paper- based documents (2) [8].
- Discontinuity of care due to lack of communication among caregivers. This is caused by distributed and fragmented medical information (3).
- Lack of evidence-based treatment due to limited access to medical records (4).
- Medical errors due to incomplete medical history or access to emergency health information (e.g. allergies, current medication list, side effects, and others) at the time of need (5). According to Sunyaev. [28], most PHRS do not offer built-in emergency access to the record, except through third-party services that are available for HealthVault. For example, Microsoft Health Vault provides users with
access codes that can be given to emergency responders and other people they trust to allow access to the emergency information.
- Limited doctor availability. For instance, patients may need to be seen on the weekend or on a holiday when their doctor’s office is closed (6).
4.2. Scenario for the Need of PHRS
John was suffering from tiredness and lack of energy for the past 4 weeks. He has several chronic conditions (type 2 diabetes, chronic kidney disease, chronic lower back pain, generalized anxiety disorder, depression, bipolar disorder, dyslipidemia, hypothyroidism, coronary artery disease and congestive heart failure) and is on multiple medications prescribed by his PCP, cardiologist, psychiatrist and pain management doctor. John had recently requested to become one of Dr. Smith’s patients. After multiple attempts, Dr. Smith was able to obtain some of his previous medical records. Dr. Smith also was interested in reviewing his previous medical diagnoses and prior/current medications. Unfortunately, there was no integration of the records and medications taken. After several interviews with John and his wife, Dr. Smith was able to determine that the patient was using the same type of long-acting insulin twice a day because one had the generic name and the other the commercial or brand name. The patient was supposed to use this insulin once a day only. This patient’s error kept his glucose at very low levels in blood which led to constant tiredness and lack of energy. Once the dose was corrected, the patient felt better. The immediate access to medical and prescription information would have allowed Dr. Smith to identify the error faster and provide him with the ability to take prompt corrective measures.
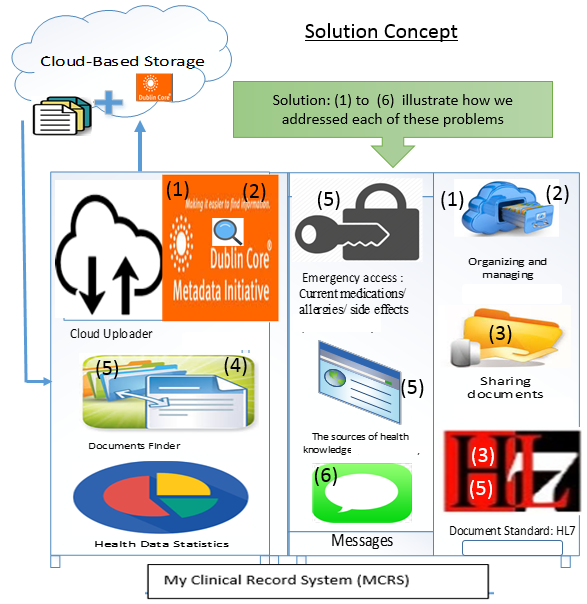
4.3. Solution Concept (TO-BE) Processes
To have continuity of care, medical records must be shared and care must be coordinated among different healthcare providers. Availability of necessary medical records could help prevent medical mistakes and enable evidence-based decisions at the point of care. It would be convenient to have clinical data stored in the same place for easy sharing and retrieval. Well-managed personal cloud space could outlive the lifetime of PHRS since clinical data is stored independently. In our approach, we separate the clinical data from applications to make the data independent from the application. Also, the users can have alternative applications to access their clinical data. Such independence helps clinical data outlive its applications. Our proposed solution concepts are illustrated in the Figure 2. In the Figure, the clinical data is separated from the application for data independence. The numbers in the Figure 2 correspond to the problems listed in the Figure 1.
5. My Clinical Record System (MCRS): A Proof of Concept
As mentioned in the introduction, there are a number of obstacles in collecting and maintaining personal health data. In an attempt to remove such obstacles, we developed a web-based system called My Clinical Record System (MCRS) that can help users to upload, organize, share, and retrieve relevant health data.
5.1. Overview of MCRS
Some of the main features of MCRS are listed below:
- Users can search their cloud storage not only by keyword, but also by any of the meta-data elements using document finder search features. They also can search by two elements such as subject and date in order to filter data by showing more relevant data. Additionally, they can find a group of records based on a date range they specify.
- MCRS allows users to share their health records with their physicians using the share document feature as shown in the Figure 3.
- MCRS helps users to create meta-data for any documents (textual and non-textual data) and upload them to their cloud storage. For the non-textual data generated from sensors such as electrocardiogram (ECG) measurement devices, the users can scan them or take a picture and then upload them as an image or use a plotted number. Thus, the user will be able to include such data into their PHR and share it with their healthcare providers. MCRS also helps to retrieve those documents easily and can direct the users to its location if more information is needed.
- To overcome the ownership barrier, we separate the clinical data from applications, which will give the users more freedom by not limiting them to one provider or application. Also, their data is saved on their own storage, thus we do not have to store it in our system.
- To overcome the interoperability barrier, we used DC standards to describe any clinical data using our tag definition. The DC meta-data content for doctor visit summary document is shown in the Figure 4 and in the Figure 5.
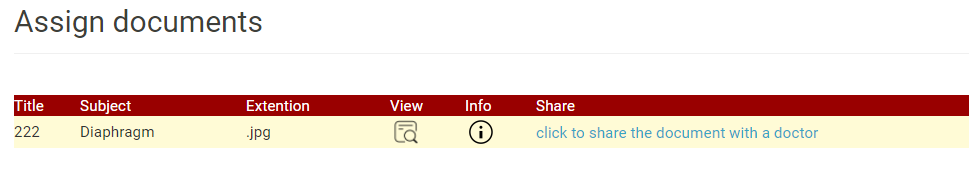 Figure 3: Sharing Health Records with Physician
Figure 3: Sharing Health Records with Physician
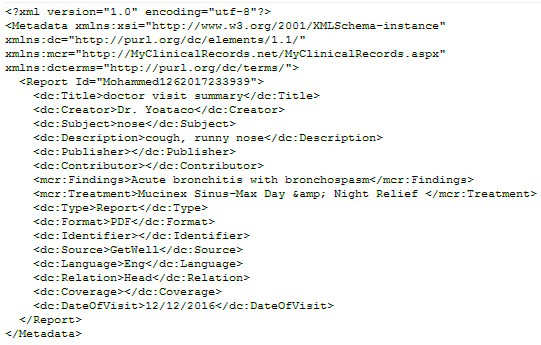 Figure 4: DC Meta-data Content for Doctor Visit Summary
Figure 4: DC Meta-data Content for Doctor Visit Summary
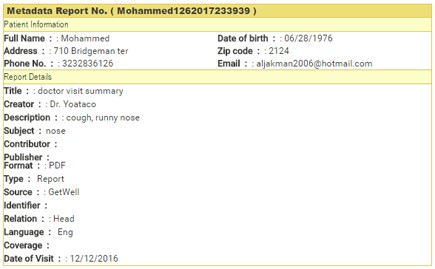 Figure 5: Visualization of the Meta-data
Figure 5: Visualization of the Meta-data
 Figure 6: Emergency Access with Valid License Number
Figure 6: Emergency Access with Valid License Number
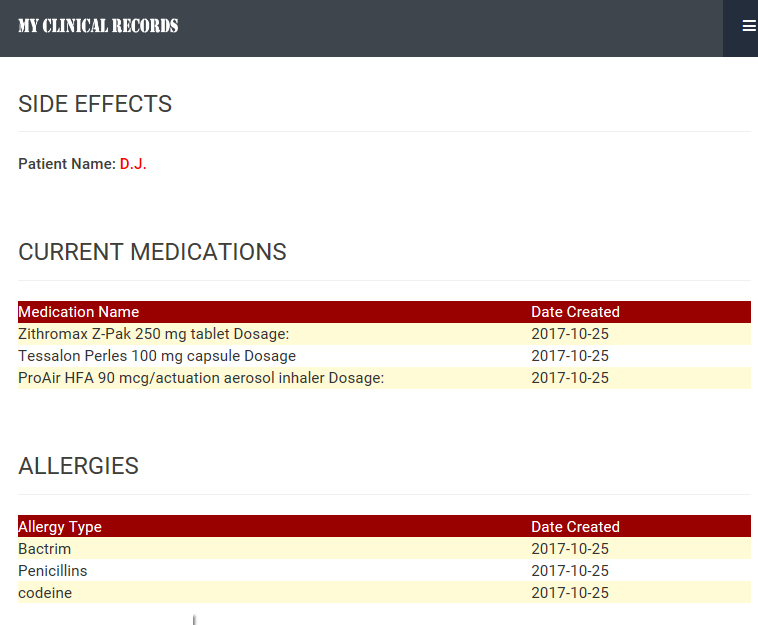 Figure 7: Emergency Information
Figure 7: Emergency Information
- The easy access to users’ health data and the ability to contribute to their record enhances users’ motivation to use PHR.
- MCRS enables emergency clinical data access by emergency crew only with valid license number. We use the National Provider Identifier (NPI), patient name, and date of birth for the emergency medical information access as shown in the Figure 6. MCRS uses the Application Programming Interface (API) that was provided by NPPES NPI Registry in order to verify the NPI. Emergency information contains allergies, current medication list, and side effects. This information is updated regularly by patients as shown in the Figure 7. It also contains any references to the time of the last update.
- Healthcare providers apply for NPI using the National Plan and Provider Enumeration System (NPPES) [29].
- NPI can be validated through NPPES NPI Registry.
5.2. Using MCRS
We use patient’s Dropbox access token to allow the connection between Dropbox and MCRS, so patients can have their own storage and have the ability to provide access to their storage through MCRS when needed. This allows users to keep their own data without binding to any specific application. MCRS contains no clinical data as they are stored in the patient’s cloud storage. Patients need accounts for Dropbox and MCRS separately.
5.3. Personal Cloud Storage
Cloud storage is a place where users can store their data and access it anytime, from anywhere, and from any device via the Internet. It is maintained, managed and operated by cloud storage service providers such as Google, Amazon, or Microsoft Cloud storage services have many advantages such as cost savings, ease of use, ability to share data, accessibility, and sustainability. Personal Cloud Storage (PCS) is getting more popular because of the aforementioned convenience. Any cloud storage service provider may be used (SugarSync, Carbonite, IDrive, Dropbox, Google Drive, and others) – for storing personal clinical data as long as they provide required functionality and security. For testing purposes, we chose Dropbox™ as cloud storage. However, for practical use, secure cloud storage services that are HIPAA complaint can be used for privacy and security purposes, such as Dropbox (Business), Box, Google Drive, Microsoft OneDrive, and Carbonite [30]. The contents of each storage are described by DC meta-data for interoperability. In the case that data has embedded meta-data, we create another layer of meta-data so entire contents can be retrieved through our DC meta-data.
5.4. Managing Health Data
MCRS categorizes clinical data based on human body parts. There are eight categories: abdomen, heart, head, thorax, extremities, integumentary, urinary, and reproductive, as shown in Table 2. Any clinical data will be stored and linked based on these categories using the relation and subject tag elements of the DC meta-data. We kept the categories to a minimum so it can be simple enough to be used by patients. Users can specify the category (the human body part of interest) when searching for relevant clinical data, so it can show only the clinical documents (e.g. doctor visit summary, x-ray, and others) that are related to that part.
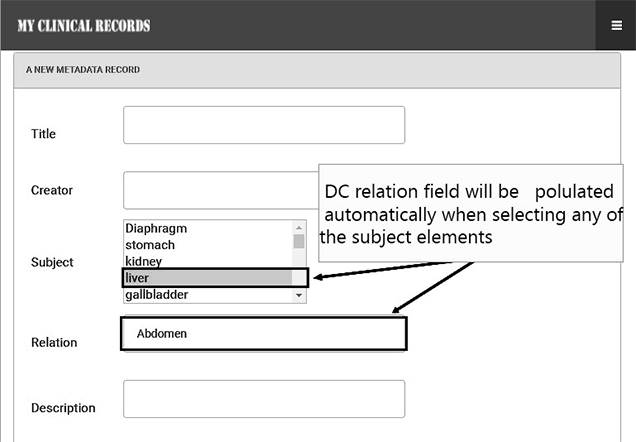
When using the relationships between the resource (DC subject) and target resource (DC relation), it is possible to combine the result to a greater scope, e.g. instead of eyes and ears, it can be categorized by head. This can be done by predefining each part of the human body and associating it with its related category in the system. Also, we have constrained the DC subject to a small core set that can be selected from a drop-down menu (all possible parts of the human body) to best describe the subjects (as shown in the Figure 8). When the users select the subject element, the DC relation field will be populated automatically with the associated part of its related category. For example, when a user searches by keyword (e.g. head) and chooses the element (e.g. relation), the search results will be filtered and show only all clinical documents that are relevant to the head (e.g. eyes, ears, brain, mouth, teeth, nose, and chin) as shown in the Figure 9. This is a less time-consuming method to filter the data instead of showing all documents as shown in the Figure 10. Also, the user can filter the search by date if they need to specify a period of time to find clinical documents.
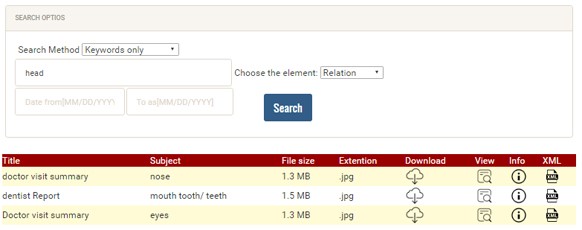 Figure 9: Example of Retrieving DC Meta-data for only Related Documents
Figure 9: Example of Retrieving DC Meta-data for only Related Documents
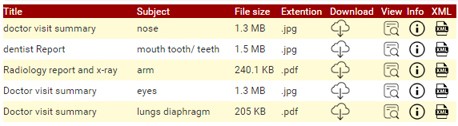 Figure 10: Example of DC Meta-data for all Documents
Figure 10: Example of DC Meta-data for all Documents
Table 2: Human Body Categories
| Body categories | Body parts |
| The abdomen | Contains diaphragm, stomach, liver, gallbladder, pancreas, small intestine, large intestine, cava, spleen, and others. |
| Heart | Contains superior vena cava, pulmonary artery, pulmonary veins, pulmonic valve, tricuspid valve, inferior vena cava, right atrium, right ventricle, left ventricle, aortic valve, mitral valve, left atrium, aorta and others. |
| Head | Contains eyes, ears, brain, mouth, teeth, nose, chin, spinal cord, tonsil, uvula, gullet, meninges, pharynx and others. |
| Thorax | Contains lungs, diaphragm/pleura, nasopharynx/oral, cavity, trachea/Larynx, ribs, capillaries, bronchial tube, windpipe/trachea, chest, esophagus and others. |
| Extremities | Contains arms, elbows, hands, wrists, shoulders, hips/thighs, fingers, thumbs, legs, knees, toe, vertebral column, neck, ankles, breast, back pain, feet and others. |
| Integumentary |
Skin and associated structures such as hair, nails, sweat glands, and oil glands
|
| Urinary |
Kidneys, ureters, urinary bladder, and urethra
|
| Reproductive |
Gonads (testes or ovaries) and associated organs; in females: uterine tubes, uterus, and vagina; in males: epididymis, ductus deferens, prostate gland, and penis
|
6. Conclusion
As the medical industry is going through a paradigm shift from clinician-centered to patient-centered, readily available complete personal medical history is becoming crucial. This will help ensure the three major goals in medical industry: evidence-based treatment, continuity of care, and prevention of medical mistakes. In this paper, we proposed an untethered personal health record system to help achieve such goals. Our proposed system, MCRS, provides a method to collect and organize heterogeneous personal health data using DC meta-data. The retrieval of the organized data was completed by the reorganized DC tags, which allowed the organization of clinical data by body parts for easy retrieval. Finally, we allowed access to personal emergency clinical data to emergency crew only by their license number at the time of need. In the future, we plan to expand the usage of clinical data collected from the application to analyze and identify the regional characteristics in health mapping to build better public policy for the nation. Our experiment was limited to one healthcare provider using the clinical data from that provider. Also, the data collection was limited due to the patient privacy regulations in the healthcare industry.
- This paper is an extension of work originally presented in 2017 IEEE/ACIS 16th International Conference on Computer and Information Science (ICIS), Wuhan, 2017.
- Key Developments in the Connected Health Markets https://www. parksassociates.com/whitepapers.
- AHIMA e-HIM Personal Health Record Work Group. “Defining the Personal Health Record.” Journal of AHIMA 76, no.6 (June 2005): 24-25.
- Richard Brandt and Rich Rice, “Building a better PHR paradigm: Lessons from the discontinuation of Google Health™”, Health Policy and Technology Volume 3, Issue 3, 2014, Pages 200-207, ISSN 2211-8837, https://doi.org/10.1016/j.hlpt.2014.04.004. (http://www.sciencedirect.com/science/article/pii/S221188371400032X)
- Morgan Price, Paule Bellwood, Nicole Kitson, Iryna Davies, Jens Weber and Francis Lau, “Conditions potentially sensitive to a Personal Health Record (PHR) intervention, a systematic review”. BMC Medical Informatics and Decision Making 15, 32 (2015), https://doi.org/10.1186/s12911-015-0159-1
- Price, Margaux M. and Pak, Richard and M{\”u}ller, Hendrik and Stronge, Aideen, “Older adults’ perceptions of usefulness of personal health records” “Universal Access in the Information Society 12, 191-204 (2013), doi=”10.1007/s10209-012-0275-y
- Alyami, M.Aalyami and Song, Yeong-Tae: Removing Barriers in using Personal Health Record Systems. 2016 IEEE/ACIS 15th International Conference on Computer and Information Science (ICIS). DOI: 10.1109/ICIS.2016.7550810
- James D. and Laitin, David D. “Ethnicity, Insurgency, and Civil War” American Political Science Review volume={97} pages={75–90}, (2003). DOI={10.1017/S0003055403000534
- Sweet Lauren E. and Moulaison Heather Lea. ” Electronic Health Records Data and Metadata: Challenges for Big Data in the United States” Big Data. January 2014, 1(4): 245-251. https://doi.org/10.1089/big.2013.0023
- Greenberg, Jane, Maria Cristina Pattuelli, Bijan Parsia, and W. Davenport Robertson. “Author-generated Dublin Core metadata for web resources: a baseline study in an organization.” In International Conference on Dublin Core and Metadata Applications, pp. 38-45. 2001.
- Talha, Norhaizan Mat. “Metadata management system (MMS).” In International Conference on Dublin Core and Metadata Applications. 2004.
- Haugen, Mary Beth and Herrin, Barry and Slivochka, Sharon and Tolley, Lori McNeil and Warner, Diana and Washington, Lydia,”Rules for handling and maintaining metadata in the EHR” Journal of AHIMA 84, no.5 (May 2013): 50-54. URL: http://europepmc.org/abstract/MED/23755436
- Dhivya and S. Roobini and A. Sindhuja, “A. Symptoms Based Treatment Based on Personal Health Record Using Cloud Computing” Procedia Computer Science 47, 22-29 (2015), doi = “https://doi.org/10.1016/j.procs.2015.03.179” url = http://www.sciencedirect.com/science/article/pii/S1877050915004470
- Chintan Patel and Karthik Gomadam and Sharib Khan and Vivek Garg, “TrialX: Using semantic technologies to match patients to relevant clinical trials based on their Personal Health Records” Web Semantics: Science, Services and Agents on the World Wide Web, 8, 342-347 (2010) doi = “https://doi.org/10.1016/j.websem.2010.08.004”, url = http://www.sciencedirect.com/science/article/pii/S1570826810000636
- Appelboom, Geoff and Camacho, Elvis and Abraham, Mickey E. and Bruce, Samuel S. and Dumont, Emmanuel LP et al. “Smart wearable body sensors for patient self-assessment and monitoring” Archives of Public Health 72, 28 (2014). doi=”10.1377/hlthaff.2012.1216″, url=https://doi.org/10.1377/hlthaff.2012.1216
- Zhang J., Chen D.& Wong T. ‘Metadata application on clinical trial data in drug development,’ Proceedings of the Twenty Eighth Annual SAS® Users Group International Conference, Seattle, WA (2003).
- Tal Teitz and Dwayne G. Stupack and Jill M. Lahti, “Halting Neuroblastoma Metastasis by Controlling Integrin-Mediated Death” Cell Cycle 5, 681-685 (2006). doi = {10.4161/cc.5.7.2615}, URL = http://dx.doi.org/10.4161/cc.5.7.2615
- Ariel Ekblaw, Asaf Azaria, Thiago Vieira, Andrew Lippman. (2016). MedRec: Medical Data Management on the Blockchain. PubPub, https://www.pubpub.org/pub/medrec version: 57e013615dbf3f3300152554].
- Asuman Dogac , Gokce B. Laleci , Yildiray Kabak , Seda Unal , Sam Heard , Thomas Beale , Peter Elkin , Farrukh Najmi , Carl Mattocks , David Webber et al. “Exploiting ebXML registry semantic constructs for handling archetype metadata in healthcare informatics” International Journal of Metadata, Semantics and Ontologies 1, 21-36 (2006). https://doi.org/10.1504/IJMSO.2006.008767
- Poeggel, Sven, Daniele Tosi, DineshBabu Duraibabu, Gabriel Leen, Deirdre McGrath, and Elfed Lewis. “Optical fibre pressure sensors in medical applications.” Sensors 15, no. 7 (2015): 17115-17148.
- Guner C, Akin S, Durna Z. Comparison of the symptoms reported by post-operative patients with cancer and nurses’ perception of patient symptoms. Eur J Cancer Care 2014;23:523-30. doi: 10.1111/ecc.12144
- Jeff Mather, MathWorks “Accessing data in DICOM files” Technical Articles and Newsletters Published 2002http://www.mathworks.com/company/newsletters/articles/accessing-data-in-dicom-files.html?requestedDomain=www.mathworks.com (accessed October 2016)
- Meltzer, Eli O., Daniel L. Hamilos, James A. Hadley, Donald C. Lanza, Bradley F. Marple, Richard A. Nicklas, Allen D. Adinoff et al. “Rhinosinusitis: developing guidance for clinical trials.” Journal of Allergy and Clinical Immunology 118, no. 5 (2006): S17-S61.
- LaConte, Grace M. “Documentation roles in an electronic health record environment.” PhD diss., The College of St. Scholastica, 2011.
- Brnson, Principles of Health Interopersability HL7 ans SNOMED CT, Health Information Technology Standards, 2012.
- Chao-chen Chen, Hsueh-hua Chen, Kuang-hua Chen. “The Design of Metadata Interchange for Chinese Information and Implementation of Metadata Management System” BULLETIN of the American Society for Information Science and Technology 27, No. 5 June / July 2001, url: https://www.asis.org/Bulletin/Jun-01/chen.html
- Informatica Metadata Management for Holistic Data Governance. (July 2013).at https://www.informatica.com/content/dam/informatica-com/global/amer/us/collateral/white-paper/metadata-management-data-governance_white-paper_2163.pdf
- Sunyaev, Ali, Dmitry Chornyi, Christian Mauro, and Helmut Krcmar. “Evaluation framework for personal health records: Microsoft HealthVault vs. Google Health.” In System Sciences (HICSS), 2010 43rd Hawaii International Conference on, pp. 1-10. IEEE, 2010.
- Bindman Using the National Provider Identifier for health care workforce evaluation. Medicare Medicaid Res Rev. 2013;3(3):1-10.
- N. Daily, “Top 20 Best Cloud Storage Providers – Reviews and Comparison of the Top Secure Solutions and Services. Find Unlimited Cloud Based Data Storage Services and Options,” 2017.